 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_3605_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 170aa MW: 18362.9 Da PI: 7.3974 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 84.2 | 3.7e-26 | 31 | 88 | 5 | 62 |
YABBY 5 ssseqvCyvqCnfCntilavsvPstslfkvvtvrCGhCtsllsvnlakasqllaaesh 62
+eq+Cyv+CnfC+t+lavsvP tslfk+vtvrCGhCt+llsvn++ +a+ +h
cra_locus_3605_iso_1_len_1119_ver_3 31 APTEQLCYVHCNFCDTVLAVSVPCTSLFKTVTVRCGHCTNLLSVNMRGLLLPTAVPNH 88
679******************************************9998877776665 PP
| |||||||
| 2 | YABBY | 78.8 | 1.7e-24 | 89 | 128 | 131 | 170 |
YABBY 131 rfikeeiqrikasnPdishreafsaaaknWahfPkihfgl 170
+++k+eiqrika+nPdishreafsaaaknWahfP+ihfgl
cra_locus_3605_iso_1_len_1119_ver_3 89 QLLKDEIQRIKAGNPDISHREAFSAAAKNWAHFPHIHFGL 128
579***********************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 5.3E-26 | 32 | 83 | IPR006780 | YABBY protein |
| Pfam | PF04690 | 7.2E-23 | 89 | 128 | IPR006780 | YABBY protein |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 170 aa Download sequence Send to blast |
MSSSSAFAPD HHHHHVQQHH LATASAAAAA APTEQLCYVH CNFCDTVLAV SVPCTSLFKT 60 VTVRCGHCTN LLSVNMRGLL LPTAVPNHQL LKDEIQRIKA GNPDISHREA FSAAAKNWAH 120 FPHIHFGLMP DQPVKKSSVC QQQQDNIGED GGVLMKDGFL GPANIGVSPY |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Promotes adaxial cell identity. Regulates the initiation of embryonic shoot apical meristem (SAM) development. {ECO:0000269|PubMed:19837869}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
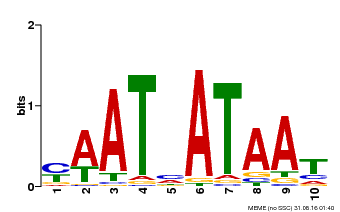
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019159560.1 | 4e-67 | PREDICTED: axial regulator YABBY 1 | ||||
| Swissprot | Q8GW46 | 2e-42 | YAB5_ARATH; Axial regulator YABBY 5 | ||||
| TrEMBL | A0A438DZZ1 | 1e-64 | A0A438DZZ1_VITVI; Axial regulator YABBY 1 | ||||
| STRING | POPTR_0002s14600.1 | 1e-68 | (Populus trichocarpa) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



