 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_3760_iso_3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 362aa MW: 39500.6 Da PI: 10.1448 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 100.2 | 1.3e-31 | 290 | 347 | 3 | 59 |
-SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 3 DgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
D y+WrKYGqK++kgs++pr+YY+C+s gCp++k+ver+ +dp+++++tYegeHnh+
cra_locus_3760_iso_3_len_1641_ver_3 290 DDYSWRKYGQKPIKGSPHPRGYYKCSSVrGCPARKHVERALDDPTMLIVTYEGEHNHS 347
99************************988****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF10533 | 1.2E-17 | 237 | 286 | IPR018872 | Zn-cluster domain |
| Gene3D | G3DSA:2.20.25.80 | 1.5E-33 | 276 | 347 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 30.733 | 283 | 349 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.92E-27 | 285 | 348 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.2E-37 | 288 | 348 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.3E-27 | 290 | 346 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 362 aa Download sequence Send to blast |
MAVELMTGFR NDGFTSKMED NGAVQEAATA GLQSVEKLIR LLSQSQSQSH QQQIYKDPSS 60 KSSATEYQAV ADAAVNKFKK FISLLDRTRT GHARFRRGPV TNPTQQQLQK PQISSPPSSS 120 SLNQPSTNLI QVQPMEEEKE KLTKSSSRIY CPTPIQGLPP LPHNHHQLVK NVPTERKESS 180 TTINFAAASP VNSFMSSLTG DTDSLQPSMT SSGFQITNLS SHVSSAGKPP LSTSSFKRKC 240 SSMDDAAVKC GGGGASSGSR CHCPKKRKSR MKRVIRVPAI SMKMADIPPD DYSWRKYGQK 300 PIKGSPHPRG YYKCSSVRGC PARKHVERAL DDPTMLIVTY EGEHNHSHSA TEAPAALVLE 360 SS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 3e-21 | 276 | 346 | 2 | 71 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
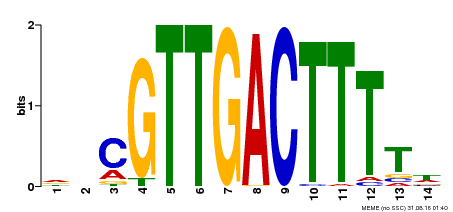
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00450 | DAP | Transfer from AT4G24240 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid and strongly during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027092449.1 | 0.0 | probable WRKY transcription factor 7 | ||||
| Swissprot | Q9STX0 | 1e-103 | WRKY7_ARATH; Probable WRKY transcription factor 7 | ||||
| TrEMBL | A0A068TYC9 | 0.0 | A0A068TYC9_COFCA; Uncharacterized protein | ||||
| STRING | XP_009604893.1 | 1e-179 | (Nicotiana tomentosiformis) | ||||



