 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_384_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 489aa MW: 56140.6 Da PI: 9.0875 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 21.3 | 6.9e-07 | 107 | 128 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C+ Cg sF+ + +L++H+++H
cra_locus_384_iso_1_len_1743_ver_3 107 VCHECGVSFKKPAYLRQHMQSH 128
6*******************99 PP
| |||||||
| 2 | zf-C2H2 | 17.2 | 1.4e-05 | 236 | 261 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp +C+++F + n++rH++ H
cra_locus_384_iso_1_len_1743_ver_3 236 FECPvdNCKRRFAFQGNMRRHMKEfH 261
89*********************988 PP
| |||||||
| 3 | zf-C2H2 | 19.4 | 2.9e-06 | 275 | 299 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y+C+ Cgk+F+ s L++H +H
cra_locus_384_iso_1_len_1743_ver_3 275 YVCSeiGCGKVFKYASKLRKHEDSH 299
99*******************9888 PP
| |||||||
| 4 | zf-C2H2 | 14.2 | 0.00013 | 366 | 390 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C +Fs+ snL++Hi+ H
cra_locus_384_iso_1_len_1743_ver_3 366 KCSfdGCAHTFSTMSNLNQHIKAvH 390
688889***************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 2.76E-6 | 60 | 129 | No hit | No description |
| SMART | SM00355 | 32 | 62 | 84 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.0E-5 | 98 | 129 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0098 | 106 | 128 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.136 | 106 | 133 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 108 | 128 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-6 | 221 | 258 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 6.75E-6 | 234 | 261 | No hit | No description |
| PROSITE profile | PS50157 | 10.221 | 236 | 266 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.005 | 236 | 261 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 238 | 261 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 7.29E-5 | 272 | 299 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.2E-7 | 272 | 299 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0015 | 275 | 299 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.427 | 275 | 304 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 277 | 299 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.63 | 307 | 332 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 309 | 332 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 16 | 335 | 356 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 1.71E-6 | 348 | 403 | No hit | No description |
| PROSITE profile | PS50157 | 10.637 | 365 | 395 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0049 | 365 | 390 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-5 | 366 | 391 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 367 | 390 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 16 | 396 | 422 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 398 | 422 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 489 aa Download sequence Send to blast |
XXWELAALAE CECALLSTPG FLTKSGAPHR NSSTSPLSEI VSKVEMVEEK SKPVIFRDIR 60 RYYCEYCGIC RSKKALISAH ILSHHQDDLK QGEEDKRETH NGPKMNVCHE CGVSFKKPAY 120 LRQHMQSHSL EAXSIGSHTK NSKMQEANVN LNVIIPGRGT SXEDPSSKPQ SRFLERRGLR 180 IFFDLMLLNK VQLNHXHAFF VLFRGLSHVQ WMTVTPAXRR KDHLTRHLLQ HQGKLFECPV 240 DNCKRRFAFQ GNMRRHMKEF HMEPSSSDEV SAKQYVCSEI GCGKVFKYAS KLRKHEDSHV 300 KLETVEAFCS EPGCMKYFTN EQCLKEHIRS CHQHITCEKC GTRQLKKNIK RHLRMHEAGV 360 SSERTKCSFD GCAHTFSTMS NLNQHIKAVH LELTPFACGF PGCGMKFSFK HVRDHHEKSS 420 RHVYSHGDFE ASDEQFRSRP RGGRKRKYPV IETLMRKRIV PPGEADPILN NGSEYLSRLL 480 SSGESEDDM |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 438 | 446 | RPRGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
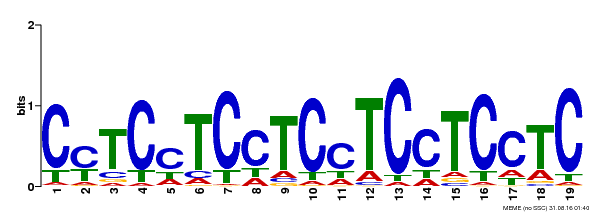
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024185714.1 | 1e-154 | transcription factor IIIA-like isoform X1 | ||||
| Swissprot | Q84MZ4 | 2e-87 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A068V6C1 | 0.0 | A0A068V6C1_COFCA; Uncharacterized protein | ||||
| STRING | VIT_08s0040g00240.t01 | 1e-173 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3644 | 24 | 47 |



