 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_384_iso_7 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 399aa MW: 46179.9 Da PI: 7.5618 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 21.7 | 5.4e-07 | 107 | 128 | 2 | 23 |
EETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirtH 23
+C+ Cg sF+ + +L++H+++H
cra_locus_384_iso_7_len_1193_ver_3 107 VCHECGVSFKKPAYLRQHMQSH 128
6*******************99 PP
| |||||||
| 2 | zf-C2H2 | 18 | 8.1e-06 | 292 | 316 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp dC+ s++rk++L+rH+ H
cra_locus_384_iso_7_len_1193_ver_3 292 FTCPvdDCHSSYRRKDHLTRHLLQH 316
89*******************9887 PP
| |||||||
| 3 | zf-C2H2 | 15 | 7.3e-05 | 321 | 346 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++Cp +C +F + n+krH++ H
cra_locus_384_iso_7_len_1193_ver_3 321 FECPgdNCQSRFAFQGNMKRHMKEfH 346
89*********************988 PP
| |||||||
| 4 | zf-C2H2 | 18.4 | 5.8e-06 | 360 | 384 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y+C+ Cgk+F+ s L++H +H
cra_locus_384_iso_7_len_1193_ver_3 360 YVCSefGCGKVFKFASKLRKHEDSH 384
99*******************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 2.82E-6 | 60 | 128 | No hit | No description |
| SMART | SM00355 | 32 | 62 | 84 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.0E-5 | 98 | 129 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0098 | 106 | 128 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.094 | 106 | 133 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 108 | 128 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-8 | 289 | 318 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 5.79E-6 | 289 | 318 | No hit | No description |
| SMART | SM00355 | 0.0059 | 292 | 316 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.64 | 292 | 321 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 294 | 316 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.75E-5 | 319 | 347 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-4 | 319 | 347 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.28 | 321 | 346 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.432 | 321 | 351 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 323 | 346 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.0E-7 | 354 | 384 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.157 | 360 | 389 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0073 | 360 | 384 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 362 | 384 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 399 aa Download sequence Send to blast |
XXWELAALAE CECALLSTPG FLTKSGAPHR NSSTSPLSEI VSKVEMVEEK SKPVIFRDIR 60 RYYCEYCGIC RSKKALISAH ILSHHQDDLK QGEEDKRETH NGPKMNVCHE CGVSFKKPAY 120 LRQHMQSHSL EXNSYSLFNF YVAIPALTST PLFFNLPNYC CLSSVRFLWF NWKIYGNKPK 180 SEEGLRLVFA LYTCDDDYPF SQTSELKTXH YVLVLYVFCI CAFNSIPXRY VPPVGSLLEA 240 ICFDDLLDIK RCNLYVXLLD FLLGVKHWIP HKELQNARGQ CXLECDQYQR PFTCPVDDCH 300 SSYRRKDHLT RHLLQHQGKL FECPGDNCQS RFAFQGNMKR HMKEFHKEPY SSDEESPKQY 360 VCSEFGCGKV FKFASKLRKH EDSHVKLETV EAFCSEPGX |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
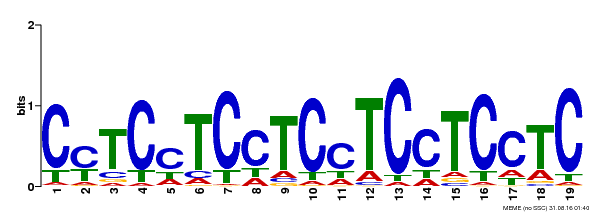
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Swissprot | Q84MZ4 | 2e-32 | TF3A_ARATH; Transcription factor IIIA | ||||
| STRING | Gorai.009G128000.1 | 1e-49 | (Gossypium raimondii) | ||||



