 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_4684_iso_3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 527aa MW: 57542.6 Da PI: 8.5857 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.7 | 4.2e-16 | 56 | 100 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+eE++++++a k++G W++I +++g +t+ q++s+ qk+
cra_locus_4684_iso_3_len_2744_ver_3 56 RERWTEEEHKKFLEALKLYGRA-WRRIEEHVG-SKTAVQIRSHAQKF 100
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.94E-16 | 50 | 104 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.107 | 51 | 105 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 5.0E-17 | 54 | 103 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 4.0E-13 | 55 | 103 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-9 | 56 | 96 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.5E-13 | 56 | 99 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.82E-10 | 58 | 101 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 527 aa Download sequence Send to blast |
MAIQGQSNNS ESSSTISIGN QISLHVDIPS TKNEQFQCED DCLPKVRKPY TITKQRERWT 60 EEEHKKFLEA LKLYGRAWRR IEEHVGSKTA VQIRSHAQKF FSKVVRESTN GDSGSGKVIE 120 IPPPRPKRKP LHPYPRKLVS PAKSGTATSQ KLTQTVSPNI SSPAEEHQSP TSVLSAPCSD 180 TPGTTDSATS DGSESLSPIS SVVGAKSVGF VLSELPDLTS ETKRSPSSSQ VNSSADNKSP 240 STSQVNNCAN QADQLHLVIA SSLILFVQIT XCDLALIYVL LNNQKLELFP QDNAFDKEGS 300 VEVSSSQIFK LFGKTVLVID PSRPSSSTPG AGKVQPSHQN DGKSNSTKVP LGESECLRRA 360 LPCKARGNLY PSSLQRECPD TLLCGPFHSL PWLMMYQGDS SASFVQVHSP IAIKAQPLLD 420 KKELHRVQSC KGGSSTGSDA ESESIETVED KNIEVKFREL SQEGEVQKQK LPLLKPGAKA 480 ASFFRQKANS SKCGKGFVPY KRCLSDRDTL SAITGEEREE QRVRLCL |
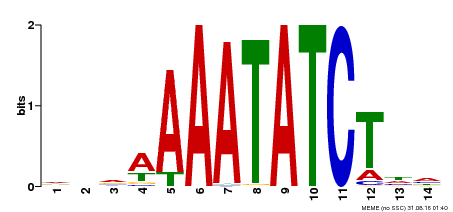
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027075006.1 | 1e-143 | protein REVEILLE 1-like isoform X1 | ||||
| TrEMBL | A1DR85 | 0.0 | A1DR85_CATRO; MYB transcription factor (Fragment) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3185 | 24 | 47 |



