 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_5090_iso_5 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | HB-other | ||||||||
| Protein Properties | Length: 207aa MW: 23270.6 Da PI: 8.81 | ||||||||
| Description | HB-other family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 62.2 | 7.8e-20 | 20 | 78 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++L +++ +++ +q+kvWFqNrR +ek+
cra_locus_5090_iso_5_len_1274_ver_3 20 KYVRYTPEQVEALERLYHECPKPSSLRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 78
5679*****************************************************97 PP
| |||||||
| 2 | bZIP_1 | 20.1 | 1.4e-06 | 72 | 117 | 18 | 63 |
HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 18 rrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
rr+R++ ++e +L+ +L+a Nk L +e+++l+k+v++l e+
cra_locus_5090_iso_5_len_1274_ver_3 72 RRCREKQRKEASRLQAVNRKLTAMNKLLMEENDRLQKQVSQLVYEN 117
9***************************************997665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-19 | 9 | 75 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 15.775 | 15 | 79 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.3E-15 | 17 | 83 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.13E-18 | 19 | 83 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 4.29E-16 | 20 | 80 | No hit | No description |
| Pfam | PF00046 | 1.9E-17 | 21 | 78 | IPR001356 | Homeobox domain |
| CDD | cd14686 | 9.22E-6 | 72 | 111 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 7.0E-4 | 76 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 207 aa Download sequence Send to blast |
MAVTSTCKDA ASKMGMDNGK YVRYTPEQVE ALERLYHECP KPSSLRRQQL IRECPILSNI 60 EPKQIKVWFQ NRRCREKQRK EASRLQAVNR KLTAMNKLLM EENDRLQKQV SQLVYENSYF 120 RQHTQNATLA TTDNSCESVV TSGQPHLTPQ HPPRDASPAG LLSIAEETLT EFLSKATGTA 180 VEWVQMPGMK PGPDSIGIIA ISHGCTG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of vascular development. May promote differentiation of precambial and cambial cells. {ECO:0000269|PubMed:11402194, ECO:0000269|PubMed:15598805}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
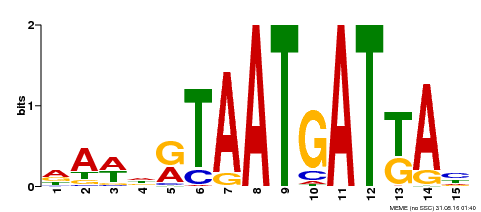
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165. {ECO:0000269|PubMed:17237362, ECO:0000269|PubMed:8575317}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011081776.1 | 1e-139 | homeobox-leucine zipper protein ATHB-15 | ||||
| Swissprot | Q39123 | 1e-125 | ATHB8_ARATH; Homeobox-leucine zipper protein ATHB-8 | ||||
| TrEMBL | A0A068UKR5 | 1e-138 | A0A068UKR5_COFCA; Uncharacterized protein | ||||
| TrEMBL | A0A2I0IVT4 | 1e-138 | A0A2I0IVT4_PUNGR; Uncharacterized protein | ||||
| STRING | XP_010047286.1 | 1e-137 | (Eucalyptus grandis) | ||||



