 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_5515_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 288aa MW: 31807.7 Da PI: 6.936 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 102 | 1e-31 | 6 | 119 | 2 | 104 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssasec.. 73
g+kdrhsk++T +g+RdRRvRls+++a++++dLqd+LG+ ++sk ++WLl +ak++i+el+ ++ +++s
cra_locus_5515_iso_2_len_1680_ver_3 6 FGGKDRHSKVCTVRGLRDRRVRLSVPTAIQLYDLQDRLGLNQPSKVVDWLLDAAKHEIDELPPLQMPPNSGNli 79
689*************************************************************7777744444 PP
TCP 74 .eaesssssasnsssg..................kaaksaakskksqksa 104
++++ + s+s+ +s+ + q+ +
cra_locus_5515_iso_2_len_1680_ver_3 80 iSSHNLQPSGSTWNSSdhiisrpkskqvsrrsddD----------HQEGS 119
44444444444333333344433322222211111..........11111 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51369 | 31.554 | 8 | 66 | IPR017887 | Transcription factor TCP subgroup |
| Pfam | PF03634 | 1.2E-30 | 8 | 128 | IPR005333 | Transcription factor, TCP |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0031347 | Biological Process | regulation of defense response | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 288 aa Download sequence Send to blast |
RVSRAFGGKD RHSKVCTVRG LRDRRVRLSV PTAIQLYDLQ DRLGLNQPSK VVDWLLDAAK 60 HEIDELPPLQ MPPNSGNLII SSHNLQPSGS TWNSSDHIIS RPKSKQVSRR SDDDHQEGSK 120 QGHDFDQDDD NHHNGTYNVS SSNNFLISNN SAHQASYTSL LRWDPSTLSL AQSGLGAAPT 180 EDHNFHNFNV VSVASAAAAA PQVLVYQPPV GITQSYFPST AEYHDHVPKH INFQMLNNSN 240 NSLTDSTSQQ QSIRPFHMAN FLSTTTQKDR AGGNELSSNN EGDFGSSR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 1e-18 | 13 | 67 | 1 | 55 | Putative transcription factor PCF6 |
| 5zkt_B | 1e-18 | 13 | 67 | 1 | 55 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00326 | DAP | Transfer from AT3G02150 | Download |
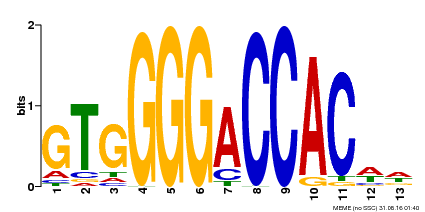
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027077547.1 | 3e-77 | transcription factor TCP5-like | ||||
| Refseq | XP_027077548.1 | 3e-77 | transcription factor TCP5-like | ||||
| TrEMBL | A0A068U1B6 | 4e-75 | A0A068U1B6_COFCA; Uncharacterized protein | ||||
| STRING | XP_009623901.1 | 7e-56 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA821 | 24 | 99 |



