 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_5840_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 405aa MW: 45794.1 Da PI: 9.6494 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 72.5 | 4.5e-23 | 19 | 84 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
rk+ksL+ll+++fl+l+ ++ +++ l+++a +L v +rRRiYDi+NVLe+++++++k+kn+++wkg
cra_locus_5840_iso_1_len_1434_ver_3 19 RKQKSLGLLCSNFLSLYDREGVETIGLDDAAARL---GV--ERRRIYDIVNVLESVGVLARKAKNRYTWKG 84
79*****************999************...**..****************************98 PP
| |||||||
| 2 | E2F_TDP | 72.2 | 6e-23 | 153 | 233 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvse.....dvknkrRRiYDilNVLealnliek.....kek 64
rkeksL lltq+f+kl+ + + ++++l+++ak L+++ ++ k+RR+YDi+NVL +++ iek ++k
cra_locus_5840_iso_1_len_1434_ver_3 153 RKEKSLALLTQNFVKLFICMDMDMISLDDAAKILLKNgkdpsMTRSKVRRLYDIANVLASMKFIEKthhpdTRK 226
79*********************************99****98889************************99** PP
E2F_TDP 65 neirwkg 71
+++rw g
cra_locus_5840_iso_1_len_1434_ver_3 227 PAFRWLG 233
*****87 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 7.4E-26 | 17 | 85 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.93E-15 | 19 | 82 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 2.2E-27 | 19 | 84 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 2.8E-20 | 20 | 84 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 3.5E-22 | 150 | 234 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 1.2E-27 | 153 | 233 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 2.0E-17 | 154 | 233 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.9E-11 | 155 | 234 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 405 aa Download sequence Send to blast |
MALNSFTPKE AGSEQIYCRK QKSLGLLCSN FLSLYDREGV ETIGLDDAAA RLGVERRRIY 60 DIVNVLESVG VLARKAKNRY TWKGFGAIPS ALQTLKDEGF KNNCSGDDGH SSARNSDDEE 120 DDRYSNATYS SQNDKQNPDP GQKPPGSSKF DNRKEKSLAL LTQNFVKLFI CMDMDMISLD 180 DAAKILLKNG KDPSMTRSKV RRLYDIANVL ASMKFIEKTH HPDTRKPAFR WLGIKGKSEI 240 CADPMALHVS KKRAFGTELT NTMIKRSKVI TKLDGDLGQA SKAQFQPQIK CEPMEDEVPR 300 SKFQVDAKTG VKSYQFGPFA PDLSPQYHNQ ALRDLFDHYV EAWKSWYVEV AEKNPVKLIS 360 XFFFLKIAEL SNNCKSATSF MTKRRWPVCT FSMPGKIPKA NFFVH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4yo2_A | 3e-37 | 19 | 225 | 4 | 228 | Transcription factor E2F8 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Inhibitor of E2F-dependent activation of gene expression. Binds specifically the E2 recognition site without interacting with DP proteins and prevents transcription activation by E2F/DP heterodimers. Controls the timing of endocycle onset and inhibits endoreduplication. {ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11867638, ECO:0000269|PubMed:15649366, ECO:0000269|PubMed:18787127}. | |||||
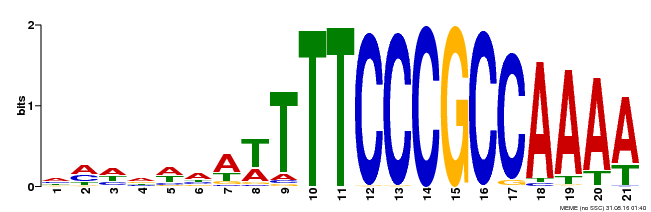
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00397 | DAP | Transfer from AT3G48160 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027066581.1 | 0.0 | E2F transcription factor-like E2FE isoform X1 | ||||
| Swissprot | Q8LSZ4 | 1e-107 | E2FE_ARATH; E2F transcription factor-like E2FE | ||||
| TrEMBL | A0A068UCS7 | 0.0 | A0A068UCS7_COFCA; Uncharacterized protein | ||||
| STRING | XP_009607525.1 | 1e-150 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2522 | 24 | 57 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



