 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_5882_iso_4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 638aa MW: 72389.5 Da PI: 5.8419 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 64.7 | 1.6e-20 | 200 | 249 | 1 | 51 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQ 51
k+r++Wt +LH++Fv+av+q+ G ek Pk+il+lm+v+ Lt+e+v+SHLQ
cra_locus_5882_iso_4_len_2744_ver_3 200 KTRVVWTVDLHQKFVKAVNQI-GFEKVGPKKILDLMGVPWLTRENVASHLQ 249
68*******************.9**************************** PP
| |||||||
| 2 | Response_reg | 82.2 | 1.7e-27 | 20 | 128 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTS CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepkl 72
vl+vdD+p+ +++l+++l+k y ev+++ ++eal+ll+e++ +D+++ D++mp+mdG++ll++ e +l
cra_locus_5882_iso_4_len_2744_ver_3 20 VLVVDDDPTWLKILEKMLKKCSY-EVTTCGLAREALDLLRERKdgFDIVISDVNMPDMDGFKLLEHVGLEM-DL 91
89*********************.***************999999**********************6644.8* PP
EEEEEESTTTHHHHHHHHHTTESEEEESS--HHHHHH CS
Response_reg 73 piivvtahgeeedalealkaGakdflsKpfdpeelvk 109
p+i+++ ge + + + ++ Ga d+l Kp+ ++el++
cra_locus_5882_iso_4_len_2744_ver_3 92 PVIMMSVDGETSRVMKGVQHGACDYLLKPIRMKELRN 128
**********************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.40.50.2300 | 1.2E-44 | 17 | 142 | No hit | No description |
| SuperFamily | SSF52172 | 1.99E-36 | 17 | 151 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 1.3E-32 | 18 | 130 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 43.773 | 19 | 134 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.6E-24 | 20 | 129 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.44E-30 | 21 | 134 | No hit | No description |
| SuperFamily | SSF46689 | 1.36E-13 | 198 | 250 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-22 | 199 | 249 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.7E-19 | 200 | 250 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.3E-6 | 202 | 249 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 638 aa Download sequence Send to blast |
MEVSGYPSPR TDAFPAGLRV LVVDDDPTWL KILEKMLKKC SYEVTTCGLA REALDLLRER 60 KDGFDIVISD VNMPDMDGFK LLEHVGLEMD LPVIMMSVDG ETSRVMKGVQ HGACDYLLKP 120 IRMKELRNIW QHVFRKRIHE VKEIENHEGF EEIQFMRNVC DQSEDRMFSS ANDLASGKKR 180 KDYENKHDDR ECSDSSSVKK TRVVWTVDLH QKFVKAVNQI GFEKVGPKKI LDLMGVPWLT 240 RENVASHLQV NICYANFSLI FHISDLCVLP PLSCVVPYMY VSVLGLLSNE RSHENVLFFC 300 LFLFQKYRLY LTRLQKEKEL TASFGVMKHS DISSKEQTGN HCLQNSVNMQ QTNGSCGISG 360 NKTAMQNIDF EIHEGDLKSL VKIPMVDANN GLGGDNPDSQ RGRRMNADHS FRKLDSDVKY 420 ATTIPEQYMW SAEGSEVQAN QERKQPFRPK PGFNPMPLPS LHQNIQFDCV RPTPSFVSKP 480 SNIHNDKLGG SEKKTIFVKS VSHGNMVDLL PVQPQSELTA TTSQEVLKPD PSSIVWSTKN 540 QRAADQIFVS DMESAQRSID LVGGWGLATW EENQQGCLIE ADIHRLTQGL QNIESFSYNE 600 AGIISEASSY LYDSLKFDYE MPLEPMEYSV IDQGLFIV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-14 | 196 | 249 | 1 | 54 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
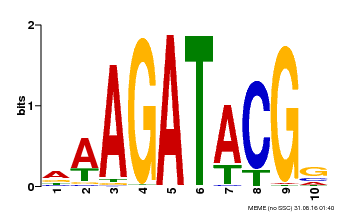
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027100212.1 | 0.0 | two-component response regulator ORR26 isoform X1 | ||||
| Refseq | XP_027152868.1 | 0.0 | two-component response regulator ORR26 | ||||
| TrEMBL | A0A2R6RED2 | 0.0 | A0A2R6RED2_ACTCH; Two-component response regulator like | ||||
| STRING | evm.model.supercontig_86.17 | 0.0 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA6788 | 24 | 33 |



