 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_6071_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 538aa MW: 61006 Da PI: 5.1692 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 75.4 | 9.1e-24 | 48 | 128 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtse 74
rW+++e+laL+++r++m+ ++r++ lk+plW+evs e gf+rs+ +Ckek+en+ k++k++k+ +++r+++
cra_locus_6071_iso_2_len_2158_ver_3 48 RWPREETLALLKIRSDMDLAFRDSTLKAPLWDEVSG---ELGFHRSARKCKEKFENIFKYHKRTKDCRSGRQNG 118
8**********************************6...78*****************************9777 PP
trihelix 75 ssstcpyfdqle 86
+s +++f+qle
cra_locus_6071_iso_2_len_2158_ver_3 119 KS--YRFFEQLE 128
76..******97 PP
| |||||||
| 2 | trihelix | 95.9 | 3.7e-30 | 359 | 443 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtse 74
rW+k ev aL+++r++++ +++++ lk+plWeevs +m++ g+ r++k+Ckekwen+nk+y+++ke++k+r +e
cra_locus_6071_iso_2_len_2158_ver_3 359 RWPKAEVEALVRLRTNLGMQFQDNGLKGPLWEEVSLAMKKLGYDRNAKRCKEKWENINKYYRRVKESQKRR-PE 431
8********************************************************************97.9* PP
trihelix 75 ssstcpyfdqle 86
ss+tcpyf+ l+
cra_locus_6071_iso_2_len_2158_ver_3 432 SSKTCPYFHLLD 443
*********987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.0035 | 45 | 104 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS50090 | 6.992 | 47 | 102 | IPR017877 | Myb-like domain |
| CDD | cd12203 | 1.37E-19 | 47 | 109 | No hit | No description |
| Pfam | PF13837 | 7.2E-15 | 47 | 129 | No hit | No description |
| SMART | SM00717 | 0.026 | 356 | 418 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS50090 | 7.422 | 358 | 416 | IPR017877 | Myb-like domain |
| Pfam | PF13837 | 1.0E-21 | 358 | 445 | No hit | No description |
| CDD | cd12203 | 5.66E-23 | 358 | 423 | No hit | No description |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 538 aa Download sequence Send to blast |
MVGESSVFLD NSGGGGDGSG GAASSAAEEM VFGGGGEEER GRGEAGNRWP REETLALLKI 60 RSDMDLAFRD STLKAPLWDE VSGELGFHRS ARKCKEKFEN IFKYHKRTKD CRSGRQNGKS 120 YRFFEQLEMF DNQPSLPSPA LNQIQSYAVE TTSATEPMVI KPTSVSLDFV ASRPTQSLNM 180 DALTPSTSTT SSSGRDSEGS IKKKRKLVDY FEKLMREVLE KQENLQNQLL NALEKCERER 240 MAREEAWKKQ QMDRIRKEQE ILAHERAIAA AKDAAVMAFL QKISEQTIPM QFPETPVPVT 300 GKHAGTDQVK TPSPLPENID KRDTVVENNI NKSDSVIEKA IEQQENGANE NFSQSSASRW 360 PKAEVEALVR LRTNLGMQFQ DNGLKGPLWE EVSLAMKKLG YDRNAKRCKE KWENINKYYR 420 RVKESQKRRP ESSKTCPYFH LLDSLYERKS NRVEQNPDWS GANLKPEDIL MQMMNRQQQQ 480 HQQPQQPQSL IEDGLRENMD QNREDEAEED DEEEDDENGN GYELVANKPS SVASMGVS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 201 | 206 | KKKRKL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00244 | DAP | Transfer from AT1G76890 | Download |
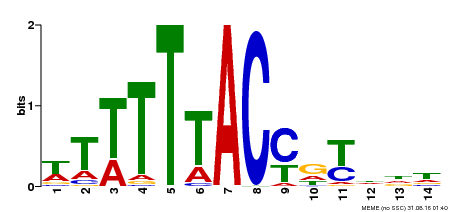
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027075443.1 | 0.0 | trihelix transcription factor GT-2-like | ||||
| TrEMBL | A0A068UW86 | 0.0 | A0A068UW86_COFCA; Uncharacterized protein | ||||
| STRING | Solyc11g005380.1.1 | 1e-162 | (Solanum lycopersicum) | ||||



