 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_7165_iso_4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 995aa MW: 112467 Da PI: 7.5609 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 160.4 | 3.4e-50 | 47 | 159 | 3 | 114 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKv 75
+e k+rwl+++ei+aiL n++ ++++ ++ + p sg+++L++rk++r+frkDG++wkkkkdgktv+E+he+LKv
cra_locus_7165_iso_4_len_3425_ver_3 47 EEaKARWLRPNEIHAILYNYKYFNVQVKPVNLPASGTMVLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKV 120
45599********************************************************************* PP
CG-1 76 ggvevlycyYahseenptfqrrcywlLeeelekivlvhy 114
g+ e +++yYah+e+nptf rrcywlL+++le+ivlv y
cra_locus_7165_iso_4_len_3425_ver_3 121 GNEERIHVYYAHGEDNPTFVRRCYWLLDKSLEHIVLVPY 159
*************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 72.074 | 42 | 168 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.5E-71 | 45 | 163 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.3E-44 | 48 | 160 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.48E-10 | 446 | 532 | IPR014756 | Immunoglobulin E-set |
| Gene3D | G3DSA:1.25.40.20 | 1.3E-17 | 628 | 743 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.99E-17 | 630 | 740 | No hit | No description |
| SuperFamily | SSF48403 | 2.64E-18 | 630 | 743 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 16.335 | 639 | 743 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.6E-6 | 654 | 743 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.487 | 681 | 713 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1.3E-4 | 681 | 710 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 270 | 720 | 749 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 430 | 792 | 816 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.035 | 830 | 856 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 61 | 845 | 867 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.346 | 846 | 875 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0019 | 868 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.834 | 869 | 893 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.8E-4 | 870 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 14 | 948 | 970 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.297 | 950 | 978 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 995 aa Download sequence Send to blast |
LKXSVGDSSR VEKEFISMDN IIAGRLVGTE IHGFRTMGDL DMANILEEAK ARWLRPNEIH 60 AILYNYKYFN VQVKPVNLPA SGTMVLFDRK MLRNFRKDGH NWKKKKDGKT VKEAHEHLKV 120 GNEERIHVYY AHGEDNPTFV RRCYWLLDKS LEHIVLVPYF XILSLDISIL LMIKFLMHFY 180 DHDIFGLKSF LIRILYQVPG THSPCSLSGN SRGAASPATP GNSNSSSGNS DPSASLVLSE 240 ESDSVVDRAY YTSHRADLVQ DESANIKDHE LRLHEINTLE WDDLLVPDDS NRRITTQEGG 300 YSVRLPYQYE TNGYRINSLP ANKLPAEDYL VNFPGLGLGN PVNNFNVPAD IGRQSMQGQI 360 TSTSLQNNFG KTALSLGNSI DNLIKDGLQT QDSFGRWIND IIIESPVSAD DMTLGSSVST 420 SHQSFTSPSM GPNLSSIPEQ YQIFYITDIS PSWASSTEPT KILVVGFFHE GLADVVKSSV 480 FCNCGDVCAP ATAIQPGVFR ILIPPQAPGV VDLFLSFDAE KPISQVVTFE FRAPRIENNL 540 ISSSEKSNWE EFQVQLRLAR LLFSTSRIFD IVSSKISPYT LKEAKTFARK TSNIADGWTR 600 LDKSIKAKEV PFPEAKDSLF ELTLQNRLYE WLLERVIEGC KITERDEQGQ GVIHLCAILG 660 YTWAVYPFSW SGLSLDYRDK LGWTALHWAA YYGREKMVAK LLSAGAKANL VSDPTSENPG 720 GCTAADLASK NGHEGLAAYL AEKALVQHFE AMTLAGNVSG SLQAPAAIDS ENPGNFTEEE 780 LNLKETLAAY RTAADAAARI QAAFREHSFK VRSKVVESSN PELEARNIIA AMKIQHAFRN 840 YETRKQMAAA ARIQYRFRTW KMRRDFLNMR RQAIKIQSVF RGFQVRRQYR KIIWSVGVLE 900 KAILRWRLKR KGFRGLQVQS DEAVKDQTQA SDTEEDFFHA SRKQAEERVE RSVVRVQAMF 960 RSKHAQEEYR RMKLEHNSAT LEYQELQNPD EMREV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
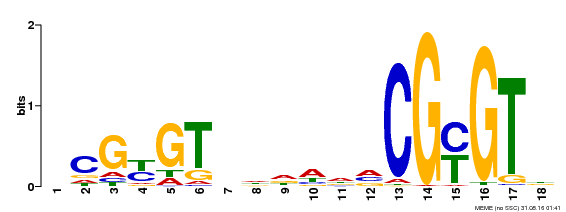
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027115917.1 | 0.0 | calmodulin-binding transcription activator 5-like | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A068TRD5 | 0.0 | A0A068TRD5_COFCA; Uncharacterized protein | ||||
| STRING | XP_009609050.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



