 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_7205_iso_3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 250aa MW: 27686 Da PI: 5.157 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 27.6 | 6.9e-09 | 12 | 38 | 20 | 48 |
TTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 20 GggtWktIartmgkgRtlkqcksrwqkyl 48
++ W++Ia++++ Rt++++k++w+++l
cra_locus_7205_iso_3_len_1000_ver_3 12 KKK-WAAIASYLP-ERTDNDIKNYWNTHL 38
566.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.447 | 1 | 42 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.48E-8 | 7 | 44 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-12 | 10 | 39 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.5E-7 | 11 | 38 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.65E-7 | 12 | 38 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0080167 | Biological Process | response to karrikin | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 250 aa Download sequence Send to blast |
XGLFLQIQGS FKKKWAAIAS YLPERTDNDI KNYWNTHLKK KLKMLQTGPD PCSRNGFSSS 60 QSASKGQWER RLQADIKTAK QALQDALTLD GTSSPVPDSK GLFSLAETGG QASNNSNTYA 120 SSTENIARLL KDWARNSSNS KSKKQSKSSS TQHSFNYPAN EYSTSSSVES KNGIELSEAF 180 ESLFGFESFD SSNSEFSQST SPEASLFQGE SKPEITQVPL AMLENWLFDE SSIQGKDLTN 240 FSFDHDEDLF |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 37 | 43 | LKKKLKM |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00395 | DAP | Transfer from AT3G47600 | Download |
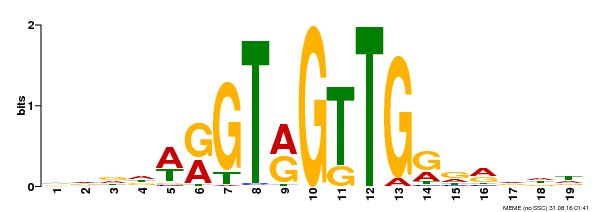
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028109092.1 | 9e-79 | myb-related protein 306-like | ||||
| Swissprot | P81392 | 2e-48 | MYB06_ANTMA; Myb-related protein 306 | ||||
| TrEMBL | A0A2N9FVI2 | 2e-76 | A0A2N9FVI2_FAGSY; Uncharacterized protein | ||||
| STRING | VIT_14s0108g00830.t01 | 4e-69 | (Vitis vinifera) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



