 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_76899_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 114aa MW: 13804.2 Da PI: 7.5176 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 68.1 | 1.1e-21 | 38 | 91 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k++++t+eq++ Le+ Fe ++++ e+++eLAkklgL+ rqV vWFqNrRa++k
cra_locus_76899_iso_1_len_340_ver_3 38 KKRRLTPEQVHLLEKSFETENKLEPERKTELAKKLGLQPRQVAVWFQNRRARWK 91
5679*************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 126.8 | 9.7e-41 | 37 | 112 | 1 | 76 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalk 74
ekkrrl+ eqv+lLE+sFe+e+kLeperK+ela++LglqprqvavWFqnrRAR+ktkqlE+dy++Lk++yd+l
cra_locus_76899_iso_1_len_340_ver_3 37 EKKRRLTPEQVHLLEKSFETENKLEPERKTELAKKLGLQPRQVAVWFQNRRARWKTKQLERDYDQLKSSYDSLL 110
69**********************************************************************98 PP
HD-ZIP_I/II 75 ee 76
++
cra_locus_76899_iso_1_len_340_ver_3 111 SN 112
76 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 18.155 | 33 | 93 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 8.4E-21 | 36 | 97 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 6.42E-21 | 38 | 104 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 6.61E-16 | 38 | 94 | No hit | No description |
| Pfam | PF00046 | 8.0E-19 | 38 | 91 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-23 | 39 | 100 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.0E-7 | 64 | 73 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 68 | 91 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.0E-7 | 73 | 89 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 2.7E-8 | 93 | 114 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001558 | Biological Process | regulation of cell growth | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 114 aa Download sequence Send to blast |
XGARSVISME ENSKRRPFFS SPEELFDEEY YDEQLPEKKR RLTPEQVHLL EKSFETENKL 60 EPERKTELAK KLGLQPRQVA VWFQNRRARW KTKQLERDYD QLKSSYDSLL SNYD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 85 | 93 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription activator involved in leaf development. Binds to the DNA sequence 5'-CAAT[AT]ATTG-3'. {ECO:0000269|PubMed:8535134}. | |||||
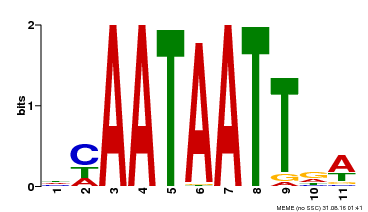
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00323 | DAP | Transfer from AT3G01470 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022873428.1 | 5e-70 | homeobox-leucine zipper protein HAT5-like | ||||
| Swissprot | Q02283 | 2e-60 | HAT5_ARATH; Homeobox-leucine zipper protein HAT5 | ||||
| TrEMBL | A0A061EJ86 | 7e-68 | A0A061EJ86_THECC; Homeobox-leucine zipper protein HAT5 | ||||
| STRING | evm.model.supercontig_215.3 | 6e-69 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA7891 | 23 | 30 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



