 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_86286_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 210aa MW: 23221.4 Da PI: 7.6073 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.9 | 6.9e-20 | 106 | 157 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++y+GVr+++ +gr++AeIrdp+++g r++lg++ t+e+Aa a+++a+ +++g
cra_locus_86286_iso_2_len_762_ver_3 106 TRYRGVRRRP-WGRFAAEIRDPNKRG--SRIWLGTYETPEDAALAYDRAAFQIRG 157
68********.**********96655..************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.5E-14 | 106 | 157 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 6.75E-29 | 106 | 167 | No hit | No description |
| SuperFamily | SSF54171 | 4.25E-23 | 107 | 166 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 6.3E-37 | 107 | 171 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 7.2E-32 | 107 | 166 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.553 | 107 | 165 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.7E-13 | 108 | 119 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 6.7E-13 | 147 | 167 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 210 aa Download sequence Send to blast |
XNDNTFLPTE VNGDHLEFLA TLTEEAGNIS PQLISVELSS EESSRISSPG DNIFISTEAN 60 RGDLGFNYVD NNSPTSTESQ EEDMAGKKKK AGAAARGKNA PTDQWTRYRG VRRRPWGRFA 120 AEIRDPNKRG SRIWLGTYET PEDAALAYDR AAFQIRGAKA LLNFPHLLNC VAQPARVGQR 180 RRAMESSSSS ESETESHGGA RKKMKQETIL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 2e-29 | 108 | 169 | 6 | 67 | ATERF1 |
| 3gcc_A | 2e-29 | 108 | 169 | 6 | 67 | ATERF1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 179 | 204 | RRRAMESSSSSESETESHGGARKKMK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. Involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways. {ECO:0000269|PubMed:11950980}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00311 | DAP | Transfer from AT2G44840 | Download |
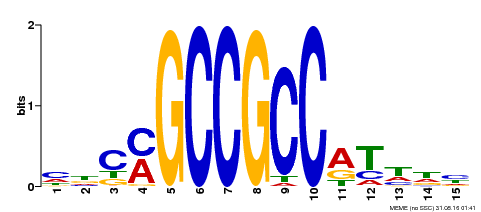
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Strongly induced by wounding. Induced by Pseudomonas syringae tomato (both virulent and avirulent avrRpt2 strains), independently of PAD4. Also induced by methyl jasmonate (MeJA) independently of JAR1, but seems to not be affected by ethylene. Induction by salicylic acid (SA) is controlled by growth and/or developmental conditions, and seems to be more efficient and independent of PAD4 in older plants. {ECO:0000269|PubMed:11950980, ECO:0000269|PubMed:12068110}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027157710.1 | 6e-43 | ethylene-responsive transcription factor 13-like | ||||
| Swissprot | Q8L9K1 | 5e-42 | ERF99_ARATH; Ethylene-responsive transcription factor 13 | ||||
| TrEMBL | A0A192XLN7 | 1e-117 | A0A192XLN7_CATRO; ORCA3-like protein 2 | ||||
| STRING | XP_009787891.1 | 3e-42 | (Nicotiana sylvestris) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



