 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_9152_iso_3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 439aa MW: 48016.2 Da PI: 6.3576 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 88.9 | 4.4e-28 | 307 | 355 | 2 | 51 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEE CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveit 51
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkkkvers+ +++++ei+
cra_locus_9152_iso_3_len_1644_ver_3 307 EDGYNWRKYGQKQVKGSEYPRSYYKCTHQNCQVKKKVERSQ-EGHITEII 355
8***************************************9.99999986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-23 | 304 | 354 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 6.28E-22 | 305 | 353 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.5E-26 | 306 | 358 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 20.3 | 307 | 347 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.8E-21 | 307 | 355 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 439 aa Download sequence Send to blast |
MGDWMPPSPS PRTLLSMLGD DVGSRSISDP IGEKIEGFSF PGPQEYMAST NADEKNGSQA 60 GLGADEAAKF SGMYEQKMSS RGGLLERRAA RAGFNAPRLN TESIRAADLL QNQEVRSPYL 120 TIPPGLSPTT LLDSPVFVNN SLVQPSPTTG KFQFVSNANS RSSTVMTEFP DKSRETFFGN 180 INPSSFAFRP MAESGPSLFF GTSNRVNPTS LPQQSFPSIE VSVQSQNVHP SSLERPNTTA 240 LNQQADFSKP STEKETGVSN NNIASEPRTL YNVGGGMDHS PPLDEQQDED ADQRSNGDPN 300 IGGAPAEDGY NWRKYGQKQV KGSEYPRSYY KCTHQNCQVK KKVERSQEGH ITEIIMALVL 360 XINQHGLSNL GLTGLGHNQI RIPSLPVHQY LGQQPRQPMN DMGLMLPKGE PKVEPVSDPG 420 LNHSNGSSVY HQVMHRMQM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 5e-13 | 308 | 356 | 16 | 62 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
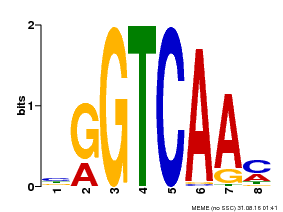
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027127652.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027127653.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027127654.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027127655.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027127656.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170840.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170841.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170842.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170844.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170845.1 | 1e-172 | probable WRKY transcription factor 2 isoform X1 | ||||
| TrEMBL | A0A336RDJ4 | 0.0 | A0A336RDJ4_CATRO; WRKY transcription factor 2 | ||||
| STRING | PGSC0003DMT400056952 | 1e-140 | (Solanum tuberosum) | ||||



