 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_9152_iso_5 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 586aa MW: 63912.9 Da PI: 6.7712 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.3 | 2.8e-32 | 163 | 220 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkkkvers+ +++++ei+Y+g Hnh+k
cra_locus_9152_iso_5_len_1957_ver_3 163 EDGYNWRKYGQKQVKGSEYPRSYYKCTHQNCQVKKKVERSQ-EGHITEIIYKGAHNHPK 220
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 104.1 | 7.5e-33 | 375 | 433 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+CtsagC+v+k+ver+++d k v++tYeg+Hnh+
cra_locus_9152_iso_5_len_1957_ver_3 375 LDDGYRWRKYGQKVVKGNPNPRSYYKCTSAGCTVRKHVERASHDLKSVITTYEGKHNHD 433
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 8.9E-29 | 160 | 221 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.55E-26 | 161 | 221 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.4E-36 | 162 | 220 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.365 | 163 | 221 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.7E-25 | 163 | 219 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 4.0E-37 | 360 | 435 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.2E-28 | 367 | 435 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.664 | 370 | 435 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.5E-37 | 375 | 434 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.9E-25 | 376 | 433 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 586 aa Download sequence Send to blast |
PSPTTGKFQF VSNANSRSST VMTEFPDKSR ETFFGNINPS SFAFRPMAES GPSLFFGTSN 60 RVNPTSLPQQ SFPSIEVSVQ SQNVHPSSLE RPNTTALNQQ ADFSKPSTEK ETGVSNNNIA 120 SEPRTLYNVG GGMDHSPPLD EQQDEDADQR SNGDPNIGGA PAEDGYNWRK YGQKQVKGSE 180 YPRSYYKCTH QNCQVKKKVE RSQEGHITEI IYKGAHNHPK PPPNRRSALG SSNALSDMQL 240 ENSEQAGTGA DGDPLWANVQ RGNGASEGRN DNPEVTSSAM MGSEYCNGAN SLQAQNGSQF 300 ETGDAVDASS TFSNDEEDDD RATHGSVSLG YDGEGDESES KRRKIETYPT DMSGASRAIR 360 EPRVVVQTTS EVDILDDGYR WRKYGQKVVK GNPNPRSYYK CTSAGCTVRK HVERASHDLK 420 SVITTYEGKH NHDVPAARNS SHVNSGGASC TLPTQPAAVQ TQVHRPEPSS QLHNSMTQFE 480 RPSSLASFAL PGRPPLGPPN HHHGFGFGIN QHGLSNLGLT GLGHNQIRIP SLPVHQYLGQ 540 QPRQPMNDMG LMLPKGEPKV EPVSDPGLNH SNGSSVYHQV MHRMQM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 8e-37 | 163 | 436 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 8e-37 | 163 | 436 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
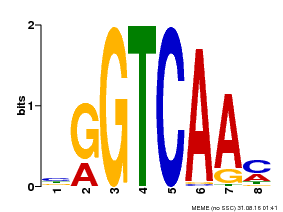
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027170840.1 | 0.0 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170841.1 | 0.0 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170842.1 | 0.0 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170844.1 | 0.0 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170845.1 | 0.0 | probable WRKY transcription factor 2 isoform X1 | ||||
| Refseq | XP_027170847.1 | 0.0 | probable WRKY transcription factor 2 isoform X3 | ||||
| Swissprot | Q9FG77 | 1e-153 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | A0A336RDJ4 | 0.0 | A0A336RDJ4_CATRO; WRKY transcription factor 2 | ||||
| STRING | VIT_19s0015g01870.t01 | 0.0 | (Vitis vinifera) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



