 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_9152_iso_8 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 684aa MW: 74362.6 Da PI: 6.7726 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102 | 3.4e-32 | 261 | 318 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkkkvers+ +++++ei+Y+g Hnh+k
cra_locus_9152_iso_8_len_2349_ver_3 261 EDGYNWRKYGQKQVKGSEYPRSYYKCTHQNCQVKKKVERSQ-EGHITEIIYKGAHNHPK 318
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 103.8 | 9.3e-33 | 473 | 531 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+CtsagC+v+k+ver+++d k v++tYeg+Hnh+
cra_locus_9152_iso_8_len_2349_ver_3 473 LDDGYRWRKYGQKVVKGNPNPRSYYKCTSAGCTVRKHVERASHDLKSVITTYEGKHNHD 531
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-28 | 258 | 319 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.18E-25 | 259 | 319 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.4E-36 | 260 | 318 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.365 | 261 | 319 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.3E-25 | 261 | 317 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.0E-37 | 458 | 533 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-28 | 465 | 533 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.664 | 468 | 533 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.5E-37 | 473 | 532 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.0E-25 | 474 | 531 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0030010 | Biological Process | establishment of cell polarity | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 684 aa Download sequence Send to blast |
MASTNADEKN GSQAGLGADE AAKFSGMYEQ KMSSRGGLLE RRAARAGFNA PRLNTESIRA 60 ADLLQNQEVR SPYLTIPPGL SPTTLLDSPV FVNNSLVQPS PTTGKFQFVS NANSRSSTVM 120 TEFPDKSRET FFGNINPSSF AFRPMAESGP SLFFGTSNRV NPTSLPQQSF PSIEVSVQSQ 180 NVHPSSLERP NTTALNQQAD FSKPSTEKET GVSNNNIASE PRTLYNVGGG MDHSPPLDEQ 240 QDEDADQRSN GDPNIGGAPA EDGYNWRKYG QKQVKGSEYP RSYYKCTHQN CQVKKKVERS 300 QEGHITEIIY KGAHNHPKPP PNRRSALGSS NALSDMQLEN SEQAGTGADG DPLWANVQRG 360 NGASEGRNDN PEVTSSAMMG SEYCNGANSL QAQNGSQFET GDAVDASSTF SNDEEDDDRA 420 THGSVSLGYD GEGDESESKR RKIETYPTDM SGASRAIREP RVVVQTTSEV DILDDGYRWR 480 KYGQKVVKGN PNPRSYYKCT SAGCTVRKHV ERASHDLKSV ITTYEGKHNH DVPAARNSSH 540 VNSGGASCTL PTQPAAVQTQ VHRPEPSSQL HNSMTQFERP SSLASFALPG RPPLGPPNHH 600 HGFGFGINQH GLSNLGLTGL GHNQIRIPSL PVHQYLGQQP RQPMNDMGLM LPKGEPKVEP 660 VSDPGLNHSN GSSVYHQVMH RMQM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-36 | 261 | 534 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-36 | 261 | 534 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Regulates WOX8 and WOX9 expression and basal cell division patterns during early embryogenesis. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Required to repolarize the zygote from a transient symmetric state. {ECO:0000269|PubMed:21316593}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00071 | PBM | Transfer from AT5G56270 | Download |
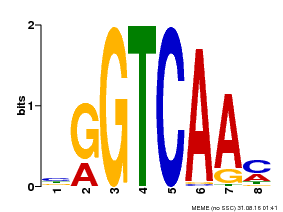
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027170847.1 | 0.0 | probable WRKY transcription factor 2 isoform X3 | ||||
| Swissprot | Q9FG77 | 0.0 | WRKY2_ARATH; Probable WRKY transcription factor 2 | ||||
| TrEMBL | A0A336RDJ4 | 0.0 | A0A336RDJ4_CATRO; WRKY transcription factor 2 | ||||
| STRING | VIT_19s0015g01870.t01 | 0.0 | (Vitis vinifera) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



