 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_9367_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 201aa MW: 22525.3 Da PI: 6.3728 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 41.9 | 1.8e-13 | 53 | 99 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
cra_locus_9367_iso_1_len_1443_ver_3 53 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAIAYITDLQ 99
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 16.806 | 49 | 98 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 4.19E-18 | 52 | 111 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.75E-13 | 52 | 103 | No hit | No description |
| Pfam | PF00010 | 6.2E-11 | 53 | 99 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-17 | 53 | 111 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.8E-16 | 55 | 104 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 201 aa Download sequence Send to blast |
MGDDEGEKLF PVRNQSVLSS LEQARDDSLI QSDERKPRKR GRKPANGRDE PLNHVEAERQ 60 RREKLNQRFY ALRAVVPNIS KMDKASLLGD AIAYITDLQK KIRTIEGEKE TSGDNNKPKN 120 IDIQDIDFHE RQEDAIVQVS CPLDAHPVSQ VVKALREHQV VAQESNVSLT ENGDVVHAFS 180 IQTPGGAAEQ LKEKLAATLL K |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 3e-31 | 47 | 109 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 34 | 42 | RKPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
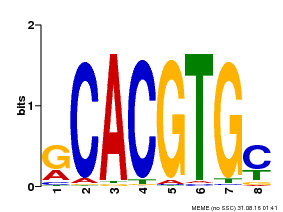
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028107222.1 | 3e-94 | transcription factor bHLH3-like | ||||
| Swissprot | A0A3Q7H216 | 3e-85 | MTB3_SOLLC; Transcription factor MTB3 | ||||
| TrEMBL | A0A2U9IY19 | 1e-139 | A0A2U9IY19_CATRO; Jasmonate-associated MYC2-like 3 | ||||
| STRING | EMJ01815 | 7e-90 | (Prunus persica) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



