 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_9526_iso_2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 247aa MW: 26396.2 Da PI: 6.3869 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 21.5 | 4.2e-07 | 3 | 31 | 22 | 55 |
HHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 22 eLrellPkaskapskKlsKaeiLekAveYIksLq 55
L+el+P++ K +Ka++L +A+eY+k Lq
cra_locus_9526_iso_2_len_1184_ver_3 3 ALQELIPNC-----NKADKASMLDEAIEYLKTLQ 31
69******8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 11.846 | 1 | 30 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.8E-10 | 1 | 39 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.36E-10 | 2 | 55 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.92E-7 | 3 | 35 | No hit | No description |
| Pfam | PF00010 | 2.1E-4 | 3 | 31 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 247 aa Download sequence Send to blast |
MRALQELIPN CNKADKASML DEAIEYLKTL QLQVQIMSMG AGLCMPPMMF PTGLQHMHGA 60 HMPHFAPMSS GMGMGFGMGM LDMNSGSPRY PIFPVPPMQG SQFPSPISGS SNFQGMAGSN 120 LPVFGHPSQG FPMSVPRTPL VPLMRQAPLS STLNLNASRM GTHAEPLRRT SPVFIHDIRP 180 QDKNSNIGQN AIASNSLSQT SAELPTTKDV DVPPASLSKD VTVQDACGNA TANLTNTDDV 240 SREEGSD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction. {ECO:0000269|PubMed:17485859}. | |||||
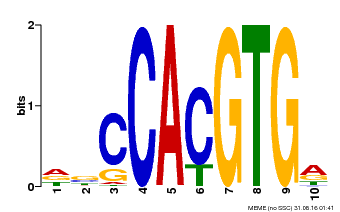
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light in dark-grown etiolated seedlings. {ECO:0000269|PubMed:17485859}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027163130.1 | 2e-92 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15-like isoform X3 | ||||
| Swissprot | Q0JNI9 | 3e-33 | PIL15_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15 | ||||
| TrEMBL | A0A0P0BZY0 | 1e-177 | A0A0P0BZY0_CATRO; Phytochrome interacting factor 3 | ||||
| STRING | PGSC0003DMT400047079 | 5e-64 | (Solanum tuberosum) | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



