 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_9629_iso_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 215aa MW: 24893.4 Da PI: 8.1009 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 96.9 | 8.4e-31 | 10 | 59 | 2 | 51 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
rien++ rqvtfskRrng+lKKA+ELSvLCda+va+iifs++gklye+ss
cra_locus_9629_iso_1_len_1032_ver_3 10 RIENETSRQVTFSKRRNGLLKKAYELSVLCDAQVALIIFSQKGKLYEFSS 59
8***********************************************96 PP
| |||||||
| 2 | K-box | 72.5 | 1.2e-24 | 80 | 171 | 8 | 98 |
K-box 8 s.leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieel 80
s + e++ ++l+ e+a++ k+ie+L+ +qR+l+G++L s+slkeL+ + +qLe+s+ iRs+K ++++e+ie+l
cra_locus_9629_iso_1_len_1032_ver_3 80 SqKVENQIQHLKYESANMAKKIEQLEASQRKLMGQNLASCSLKELEDIDDQLERSIGIIRSRKVQIYKEEIERL 153
2367888******************************************************************* PP
K-box 81 qkkekelqeenkaLrkkl 98
+ ke l een +L +k
cra_locus_9629_iso_1_len_1032_ver_3 154 KAKEILLLEENARLCHKC 171
**************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 32.66 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 5.1E-42 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 6.8E-34 | 3 | 81 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.66E-42 | 3 | 78 | No hit | No description |
| PRINTS | PR00404 | 1.3E-31 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.1E-27 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.3E-31 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.3E-31 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 6.5E-24 | 84 | 172 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.65 | 87 | 177 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009838 | Biological Process | abscission | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0080187 | Biological Process | floral organ senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 215 aa Download sequence Send to blast |
MVRGKIQMRR IENETSRQVT FSKRRNGLLK KAYELSVLCD AQVALIIFSQ KGKLYEFSSS 60 NMQRTIERYR ECAKEQQIDS QKVENQIQHL KYESANMAKK IEQLEASQRK LMGQNLASCS 120 LKELEDIDDQ LERSIGIIRS RKVQIYKEEI ERLKAKEILL LEENARLCHK CELKEPENPP 180 TSKVMNETAT CSQSTTDQSS EVETELYIGL PESRG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 2e-21 | 1 | 71 | 1 | 71 | MEF2C |
| 5f28_B | 2e-21 | 1 | 71 | 1 | 71 | MEF2C |
| 5f28_C | 2e-21 | 1 | 71 | 1 | 71 | MEF2C |
| 5f28_D | 2e-21 | 1 | 71 | 1 | 71 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | MADS-box transcription factor that acts with AGL71 and AGL72 in the control of flowering time. Promotes flowering at the shoot apical and axillary meristems. Seems to act through a gibberellin-dependent pathway. Interacts genetically with SOC1 and its expression is directly regulated by SOC1 (PubMed:21609362). Plays a role in controlling flower organ senescence and abscission by repressing ethylene responses and regulating the expression of BOP2 and IDA (PubMed:21689171). {ECO:0000269|PubMed:21609362, ECO:0000269|PubMed:21689171}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00576 | DAP | Transfer from AT5G62165 | Download |
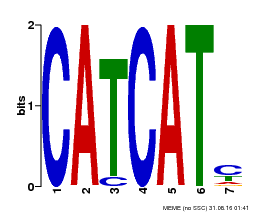
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027103881.1 | 4e-99 | MADS-box protein AGL42-like | ||||
| Swissprot | Q9FIS1 | 7e-78 | AGL42_ARATH; MADS-box protein AGL42 | ||||
| TrEMBL | A0A068UCB2 | 9e-98 | A0A068UCB2_COFCA; Uncharacterized protein | ||||
| STRING | XP_002510866.1 | 1e-84 | (Ricinus communis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |



