 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | cra_locus_9787_iso_4 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Gentianales; Apocynaceae; Rauvolfioideae; Vinceae; Catharanthinae; Catharanthus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 217aa MW: 24311.4 Da PI: 11.2949 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 99 | 3.4e-31 | 148 | 201 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHL +YR+
cra_locus_9787_iso_4_len_1761_ver_3 148 PRMRWTTTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLRMYRT 201
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 9.5E-15 | 145 | 201 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-27 | 146 | 201 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.7E-22 | 148 | 201 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.7E-7 | 149 | 200 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 217 aa Download sequence Send to blast |
MAKSDSNSFE LSLANNPNRV SSNSDHPNIN ISNLIQSTNS SNLIHSFPHP HHQIHHHHLL 60 HHHHQQNNNN NQAGLMSSEL GFLRPIRGIP VYQNHLSSSS LDKLIVPSTT TSNTFSSTTT 120 PAASFPSQNL MRSRILSRFP AKRSMRAPRM RWTTTLHARF VHAVELLGGH ERATPKSVLE 180 LMDVKDLTLA HVKSHLRMYR TVKTTDRAAA SSGNVCY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-16 | 149 | 203 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
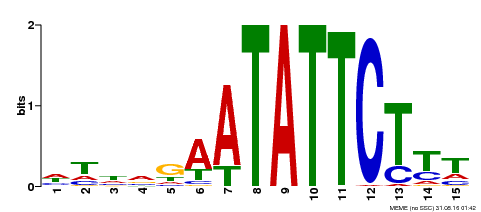
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022756946.1 | 4e-72 | probable transcription factor KAN2 isoform X2 | ||||
| TrEMBL | A0A067KUB8 | 4e-69 | A0A067KUB8_JATCU; Uncharacterized protein | ||||
| TrEMBL | B9RZ09 | 8e-70 | B9RZ09_RICCO; Transcription factor, putative | ||||
| STRING | XP_002518978.1 | 1e-70 | (Ricinus communis) | ||||



