 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | e_gw1.09.00.361.1 | ||||||||
| Common Name | OT_ostta09g00450 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Bathycoccaceae; Ostreococcus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 170aa MW: 19812.4 Da PI: 10.153 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59.8 | 5.7e-19 | 8 | 55 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEdell +av+ + g++Wk+Ia ++ +Rt+ qc +rwqk+l
e_gw1.09.00.361.1 8 KGGWTPEEDELLRRAVAVHNGKNWKKIAVHFSDTRTDVQCLHRWQKVL 55
688******************************************986 PP
| |||||||
| 2 | Myb_DNA-binding | 68.2 | 1.4e-21 | 61 | 107 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WTteEd +++++v++lG+++W++Ia++++ gR +kqc++rw+++l
e_gw1.09.00.361.1 61 KGPWTTEEDSKIIQLVTELGPKRWSKIASELP-GRIGKQCRERWYNHL 107
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 54.9 | 2.1e-17 | 113 | 157 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r W+ eEd +l+ a++++G++ W+ Ia+ + gRt++ +k++w++
e_gw1.09.00.361.1 113 REEWSSEEDRQLILAHAEYGNR-WAEIAKSFK-GRTDNAIKNHWNST 157
789*******************.********9.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.689 | 1 | 55 | IPR017930 | Myb domain |
| SMART | SM00717 | 9.3E-16 | 7 | 57 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-16 | 8 | 55 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.24E-17 | 9 | 65 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.5E-25 | 10 | 75 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.32E-11 | 11 | 55 | No hit | No description |
| PROSITE profile | PS51294 | 31.518 | 56 | 111 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.27E-31 | 58 | 154 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.5E-18 | 60 | 109 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.5E-19 | 61 | 107 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.30E-14 | 63 | 107 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-28 | 76 | 115 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 22.484 | 112 | 162 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.6E-17 | 112 | 160 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.7E-16 | 113 | 157 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.52E-11 | 115 | 158 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 7.1E-23 | 116 | 162 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 170 aa Download sequence Send to blast |
GPTRRSAKGG WTPEEDELLR RAVAVHNGKN WKKIAVHFSD TRTDVQCLHR WQKVLNPDLV 60 KGPWTTEEDS KIIQLVTELG PKRWSKIASE LPGRIGKQCR ERWYNHLDPE IKREEWSSEE 120 DRQLILAHAE YGNRWAEIAK SFKGRTDNAI KNHWNSTLKR KVDQALNQGL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 4e-67 | 7 | 162 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 4e-67 | 7 | 162 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in abiotic stress responses (PubMed:17293435, PubMed:19279197). May play a regulatory role in tolerance to salt, cold, and drought stresses (PubMed:17293435). Transcriptional activator that binds specifically to a mitosis-specific activator cis-element 5'-(T/C)C(T/C)AACGG(T/C)(T/C)A-3', found in promoters of cyclin genes such as CYCB1-1 and KNOLLE (AC Q84R43). Positively regulates a subset of G2/M phase-specific genes, including CYCB1-1, CYCB2-1, CYCB2-2, and CDC20.1 in response to cold treatment (PubMed:19279197). {ECO:0000269|PubMed:17293435, ECO:0000269|PubMed:19279197}. | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
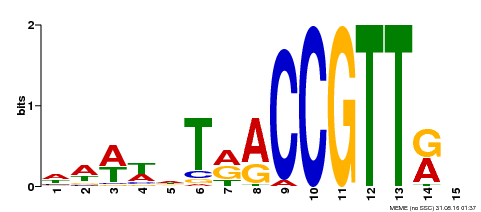
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by cold, drought and salt stresses. {ECO:0000269|PubMed:17293435}. | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022839549.1 | 1e-123 | SANT/Myb domain | ||||
| Swissprot | Q0JHU7 | 1e-75 | MB3R2_ORYSJ; Transcription factor MYB3R-2 | ||||
| Swissprot | Q6R032 | 7e-76 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | A0A090N3Y5 | 1e-122 | A0A090N3Y5_OSTTA; SANT/Myb domain | ||||
| TrEMBL | A0A454XLT8 | 1e-122 | A0A454XLT8_OSTTA; Transcription factor myb | ||||
| STRING | A0A090N3Y5 | 1e-123 | (Ostreococcus tauri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 3e-78 | myb domain protein 3r-5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



