 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | e_gw1.4.453.1 | ||||||||
| Common Name | CHLNCDRAFT_20303 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Chlorellales; Chlorellaceae; Chlorella
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 100aa MW: 11441.1 Da PI: 10.6229 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.2 | 9.3e-14 | 1 | 40 | 7 | 47 |
HHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 7 eEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+Ed+ll +++k++G+++W+ Ia+ + gR++k+c++rw +
e_gw1.4.453.1 1 QEDDLLRQLIKEYGPKNWSIIANGIK-GRSGKSCRLRWCNQ 40
69***********************9.***********985 PP
| |||||||
| 2 | Myb_DNA-binding | 50 | 6.9e-16 | 47 | 91 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+++++++Ed ++ a++++G+ W++I++ + gRt++ +k++w++
e_gw1.4.453.1 47 KDPFSQWEDAVIIMAHQKHGNA-WARISKLLV-GRTDNAVKNHWNST 91
689*******************.*********.***********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.54E-25 | 1 | 88 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 24.417 | 1 | 45 | IPR017930 | Myb domain |
| CDD | cd00167 | 6.26E-11 | 1 | 39 | No hit | No description |
| Pfam | PF13921 | 2.7E-14 | 1 | 55 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 6.2E-21 | 1 | 48 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-7 | 1 | 43 | IPR001005 | SANT/Myb domain |
| SMART | SM00717 | 1.3E-13 | 46 | 94 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.323 | 46 | 96 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-22 | 49 | 95 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.16E-8 | 49 | 92 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 100 aa Download sequence Send to blast |
QEDDLLRQLI KEYGPKNWSI IANGIKGRSG KSCRLRWCNQ LNPEVKKDPF SQWEDAVIIM 60 AHQKHGNAWA RISKLLVGRT DNAVKNHWNS TLKRKYQGG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 2e-36 | 1 | 95 | 10 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 2e-36 | 1 | 95 | 10 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 2e-36 | 1 | 95 | 10 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in auxin response. Functions in auxin signal transduction and modulates lateral root growth. Interacts with ARF response factors to promote auxin-responsive gene expression (PubMed:17675404). In response to auxin, binds sequence-specific motifs in the promoter of the auxin-responsive gene IAA19, and activates IAA19 transcription. The IAA19 transcription activation by MYB77 is enhanced by direct interaction between MYB77 and PYL8 (PubMed:24894996). {ECO:0000269|PubMed:17675404, ECO:0000269|PubMed:24894996}. | |||||
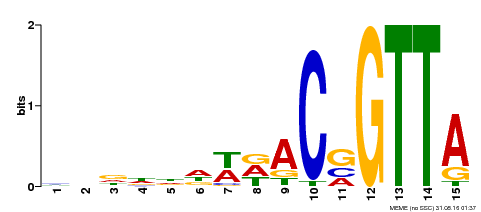
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00400 | DAP | Transfer from AT3G50060 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by auxin (PubMed:17675404). Down-regulated by potassium deprivation (PubMed:15173595). {ECO:0000269|PubMed:15173595, ECO:0000269|PubMed:17675404}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_005850232.1 | 1e-68 | hypothetical protein CHLNCDRAFT_20303, partial | ||||
| Swissprot | Q9SN12 | 2e-43 | MYB77_ARATH; Transcription factor MYB77 | ||||
| TrEMBL | E1Z785 | 3e-67 | E1Z785_CHLVA; Uncharacterized protein (Fragment) | ||||
| STRING | XP_005850232.1 | 4e-68 | (Chlorella variabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G50060.1 | 7e-46 | myb domain protein 77 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 17357546 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



