 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | estExt_fgenesh1_pg.C_60324 | ||||||||
| Common Name | COCSUDRAFT_47211 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Trebouxiophyceae incertae sedis; Coccomyxaceae; Coccomyxa; Coccomyxa subellipsoidea
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 236aa MW: 25315.9 Da PI: 10.6127 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 44.6 | 3.3e-14 | 36 | 82 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
++WT+ E+++++ + k+lG g+W+ I+r + +Rt+ q+ s+ qkyl
estExt_fgenesh1_pg.C_60324 36 QPWTEAEHLQFLTGLKKLGRGNWRGISRLFVPTRTPTQVASHAQKYL 82
79*****************************9**************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.85 | 30 | 86 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.27E-15 | 33 | 86 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-11 | 34 | 82 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.2E-10 | 34 | 84 | IPR001005 | SANT/Myb domain |
| TIGRFAMs | TIGR01557 | 3.0E-14 | 35 | 85 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.0E-11 | 36 | 82 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.74E-11 | 37 | 82 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000122 | Biological Process | negative regulation of transcription from RNA polymerase II promoter | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0030307 | Biological Process | positive regulation of cell growth | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:2000469 | Biological Process | negative regulation of peroxidase activity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 236 aa Download sequence Send to blast |
MAFELPPRLR LEERKVEVAK ANNSRDSPAG NSRKGQPWTE AEHLQFLTGL KKLGRGNWRG 60 ISRLFVPTRT PTQVASHAQK YLLRQTTVSK RKSRFCLLEQ AASAQGLLCT GVADSCGAGP 120 AMLPVFYGVQ TYPVGQLSDR AGSLQGGSAS SSDTSERGCH RMLDVPSSSC PESSRAPKFT 180 KICRPVPYHA SARLTDMAKL RADLAAAVAD DKLGPLQASS RSAFFPVRPS SAISV* |
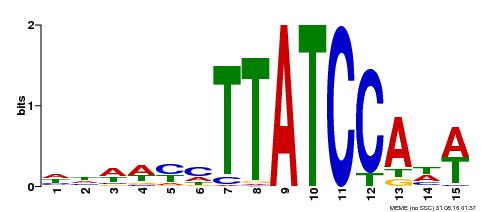
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00551 | DAP | Transfer from AT5G47390 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_005648808.1 | 1e-172 | hypothetical protein COCSUDRAFT_47211 | ||||
| TrEMBL | I0Z0U5 | 1e-171 | I0Z0U5_COCSC; Uncharacterized protein | ||||
| STRING | XP_005648808.1 | 1e-172 | (Coccomyxa subellipsoidea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP322 | 15 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47390.1 | 3e-27 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 17042262 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



