 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm.model.supercontig_19.148 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Caricaceae; Carica
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 511aa MW: 57513.6 Da PI: 7.4444 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.3 | 6.4e-32 | 107 | 160 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH +Fv+ave+LGG+e+AtPk +l+lm++kgL+++hvkSHLQ+YR+
evm.model.supercontig_19.148 107 PRLRWTPELHLCFVHAVERLGGQERATPKLVLQLMNIKGLSIAHVKSHLQMYRS 160
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.3E-30 | 103 | 161 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 13.012 | 103 | 163 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.16E-14 | 107 | 161 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-22 | 107 | 162 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.8E-9 | 108 | 159 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 511 aa Download sequence Send to blast |
MKSTHHHHHH HTTTGSSSQI SKTSNHNNFP FDNLSPKKDS EDDDDQDQEL DRGEADRIIH 60 GPSSSNSSSS VDEEKEINGG LKKTKNPGEG GSGSSSSVRQ YVRSNMPRLR WTPELHLCFV 120 HAVERLGGQE RATPKLVLQL MNIKGLSIAH VKSHLQMYRS KKISEYPNQA TTDQGFLFEG 180 GDSHVYNLSQ LSMLQNFNPR PSSSSTLRYS DPSWRNIHGV QNQIDSPNYS YKVGTSLMDR 240 ARLGFGNGAN LVLPAQKIYG NNKTNYNSHL GNSWFNSEAD RKFRRSRSEI SLETFRSFGR 300 EDKTEPESLF ESRNASARSC FRVQRFPSFN RISEDQNLPT SLTKHSENQL IINDQRQAGH 360 NYDHQAAIVT NYLENYNSRS SYMINNTNSW PSSLKQLRIQ ERQNLFSKRK ASSDSGSDAL 420 DLNLSLKVTE NINDDQNNNH GDSDESNNDD RVIDSTLSLS LSSSSSSKKR LHNRSLKEGD 480 DDDGNDEDNG KMKIHTRVAS RSSTDLDLTL * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 2e-18 | 108 | 162 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-18 | 108 | 162 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-18 | 108 | 162 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-18 | 108 | 162 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
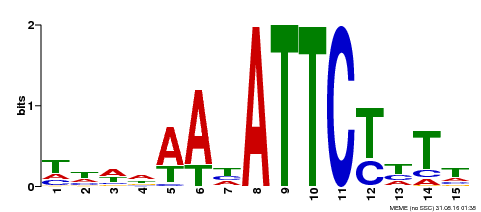
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | evm.model.supercontig_19.148 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021904128.1 | 0.0 | uncharacterized protein LOC110819301 isoform X1 | ||||
| STRING | evm.model.supercontig_19.148 | 0.0 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM363 | 28 | 184 | Representative plant | OGRP285 | 15 | 123 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 1e-45 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm.model.supercontig_19.148 |



