 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm.model.supercontig_33.151 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Caricaceae; Carica
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 559aa MW: 60609.5 Da PI: 5.6674 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.1 | 4.1e-18 | 37 | 84 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ed +l+d+vk++G g+W+++ ++ g+ R++k+c++rw ++l
evm.model.supercontig_33.151 37 KGPWTSAEDAILIDYVKKHGEGNWNAVQKHSGLSRCGKSCRLRWANHL 84
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 52.5 | 1.1e-16 | 90 | 133 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++++eE++l++++++++G++ W++ a++++ gRt++++k++w++
evm.model.supercontig_33.151 90 KGAFSQEEEQLIIELHAKMGNK-WARMAAHLP-GRTDNEIKNYWNT 133
799*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.113 | 32 | 84 | IPR017930 | Myb domain |
| SMART | SM00717 | 7.0E-16 | 36 | 86 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.42E-30 | 36 | 131 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 6.5E-16 | 37 | 84 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-24 | 38 | 91 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.31E-12 | 39 | 84 | No hit | No description |
| PROSITE profile | PS51294 | 26.43 | 85 | 139 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.7E-17 | 89 | 137 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.8E-16 | 90 | 133 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.4E-26 | 92 | 138 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.33E-12 | 92 | 133 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0045926 | Biological Process | negative regulation of growth | ||||
| GO:0048235 | Biological Process | pollen sperm cell differentiation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 559 aa Download sequence Send to blast |
MSHTTNESDE GILPKDQIFT DEGNSGGSAG GGIVLKKGPW TSAEDAILID YVKKHGEGNW 60 NAVQKHSGLS RCGKSCRLRW ANHLRPNLKK GAFSQEEEQL IIELHAKMGN KWARMAAHLP 120 GRTDNEIKNY WNTRIKRRQR AGLPLYPPEM CLQAFKESQQ GQNASGINSG DKGHHDLLQA 180 NSFEIPDVIF DSLKGSHSVL PYVPELPGMS NSSMLMKGFS SSQYCSFLPP STHRQKRIRE 240 STSLLAAYGD SVKDDFSSFN SFHDNIHQKV ARSFGLSFPH DPDPNAKTPD AFGVDQGGHT 300 LSNGNFSTSR PILEAMKLEL PSLQYPETDL GSWSRSCSPP ALLEPVDAFI QSPPRLGTDE 360 SDCVSPRNSG LLDALLHEAK NISSTKNHSH SSEKVCSSTI LADMGDSSTP PMCCDPKWGE 420 YGDPLSPLGH STTSLFSDCA PFSASGSSLD ESPPAEFTGC KLKSESVDLA WTPEGKEEPT 480 RFNITQPDIL LASDWLEQNS GHVKDPGMVS DALANLLGDD LGNEYKQMAA GGSSGQGWGL 540 GSCAWNNMPA VCQMTELP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 9e-32 | 35 | 138 | 5 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00490 | DAP | Transfer from AT5G06100 | Download |
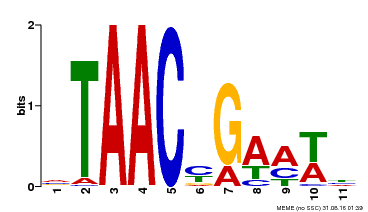
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | evm.model.supercontig_33.151 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021901215.1 | 0.0 | transcription factor MYB33 | ||||
| Refseq | XP_021901216.1 | 0.0 | transcription factor MYB33 | ||||
| TrEMBL | A0A067L7W6 | 0.0 | A0A067L7W6_JATCU; MYB family protein | ||||
| STRING | evm.model.supercontig_33.151 | 0.0 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3802 | 28 | 59 | Representative plant | OGRP5 | 17 | 1784 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06100.3 | 1e-133 | myb domain protein 33 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm.model.supercontig_33.151 |



