 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm.model.supercontig_36.105 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Caricaceae; Carica
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 951aa MW: 105686 Da PI: 7.3626 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.8 | 9.9e-41 | 165 | 241 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
Cqv++C++dls+ak+yhrrhkvCe hska+++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+q+
evm.model.supercontig_36.105 165 CQVDNCKEDLSNAKDYHRRHKVCELHSKATKALVGKQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQP 241
**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.2E-33 | 159 | 226 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.069 | 162 | 239 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.62E-37 | 163 | 243 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.2E-30 | 165 | 238 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 951 aa Download sequence Send to blast |
MEEVGAQVAP PIFIHQTLTS RFCEAPPIGR KRDLSYQTPN FLPHHNHQQQ LLPPSNSQQR 60 FMPARDNWNP KLWDWDSVRF VAQPLDPEVL RLGNPAATEQ RKIKDYIGGN GNAALTLKKN 120 AVGPEEDESL QLNLGGGLVA VEEPVSRSNK RVRSRSPGGG NYPKCQVDNC KEDLSNAKDY 180 HRRHKVCELH SKATKALVGK QMQRFCQQCS RFHPLSEFDE GKRSCRRRLA GHNRRRRKTQ 240 PEDVTSRMLL PGSQDNTTNG KLDLVNLLAA FARAQGKNED KSVNSSTVPD RDQLINILNK 300 INSLPLPVDL ASKLQNVGGF TKKLPEQPLS NHQNEMNGTS RSTMDLLAVL SATLAASSPD 360 ALALLSQKSS QSTVSDKTMS TVPDQTAGTN LQKCLEFPAV GGERSSSSYQ SPVDDSDGQV 420 QDGRSNLPLQ LFCSSPEDDS PPKLPSSRKY FSSDSSNPIE ERSPSSSPVV QKLFPMQVTT 480 GTIMSEKMSI DKEGKTHVES SRTRGMPLEL FGRPNRGADN GSVQSFPYQA GYTSSSGSDH 540 SPSSLNSDAQ DRTGRISFKL FDKDPSHFPA ELRTQIFNWL SNSPSEMESY IRPGCVVLSV 600 YVLMSSAAWE QLEGNLLQQV NCLVQDSYSD FWREARFLVH IGRRLASHKD GKIRLCKSWR 660 TWSSPELILV SPLAVVGGQE TTLSLRGRNL SNQRTKIHCM YTGGYKTMEI TGSTCQGTAY 720 DEIKLGGFRI QDACPGVLGR CFIEVENGFK GNSFPVIIAD ATICNELRVL ENEFDDDTKV 780 CDIISEEKAR DFDRPISREE ILHFLNELGW LFQRKATITK LESSDYSLSR FKFLLVFSVE 840 RDFCALVKAL IDMLVESNLD IHGLSKESLE TLTEIQLLNR AVKRRSRKMA DLLIHYSIGV 900 ADDTSRKYIF PPNLVGPGGI TPLHLAACTS DSDDMIDALT NDPQEFVSSE * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-29 | 156 | 238 | 2 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 879 | 884 | RAVKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
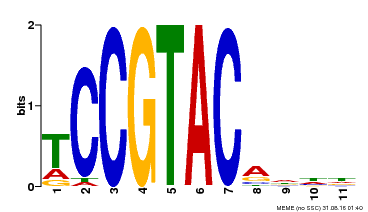
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | evm.model.supercontig_36.105 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021900760.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A061FG27 | 0.0 | A0A061FG27_THECC; Squamosa promoter binding protein-like 14 | ||||
| STRING | evm.model.supercontig_36.105 | 0.0 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 | Representative plant | OGRP7138 | 11 | 15 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm.model.supercontig_36.105 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



