 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm.model.supercontig_660.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Caricaceae; Carica
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 244aa MW: 28103.1 Da PI: 8.4419 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 97.6 | 5.3e-31 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krienk+nrqvtf+kRrng+lKKA+ELSvLCdae+a+iifs +gklye++s
evm.model.supercontig_660.1 9 KRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEIALIIFSARGKLYEFCS 59
79***********************************************96 PP
| |||||||
| 2 | K-box | 93.6 | 3.4e-31 | 82 | 172 | 9 | 99 |
K-box 9 leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqee 90
+++++ ++ +q++ kLk+++e Lq++qRhllGe+L++L keL+qLe+qL+ sl+++Rs+K++++++q++el+kke++l ++
evm.model.supercontig_660.1 82 QTARDKQNNYQDYLKLKSKVEILQHSQRHLLGEELGDLGTKELEQLEHQLDMSLRQVRSTKTQFMIDQLSELRKKEEMLMDT 163
5677889*************************************************************************** PP
K-box 91 nkaLrkkle 99
n+ Lr+kle
evm.model.supercontig_660.1 164 NRVLRRKLE 172
*******98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 33.403 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 5.4E-42 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.05E-44 | 2 | 78 | No hit | No description |
| SuperFamily | SSF55455 | 7.33E-34 | 2 | 83 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.1E-32 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 8.8E-26 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.1E-32 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.1E-32 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.3E-27 | 82 | 171 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 15.367 | 87 | 177 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010076 | Biological Process | maintenance of floral meristem identity | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048441 | Biological Process | petal development | ||||
| GO:0048442 | Biological Process | sepal development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 244 aa Download sequence Send to blast |
MGRGKVELKR IENKINRQVT FAKRRNGLLK KAYELSVLCD AEIALIIFSA RGKLYEFCSS 60 SSMVKTLERY QRVSYEALEA TQTARDKQNN YQDYLKLKSK VEILQHSQRH LLGEELGDLG 120 TKELEQLEHQ LDMSLRQVRS TKTQFMIDQL SELRKKEEML MDTNRVLRRK LEASDAALQP 180 HSWETTDHNI PFSRELTNPD AFYQPLQCNN TLQIGYNPAV TADEMIIASS SQNINGFIPG 240 WML* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 4e-25 | 92 | 171 | 22 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 4e-25 | 92 | 171 | 22 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 4e-25 | 92 | 171 | 22 | 101 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 4e-25 | 92 | 171 | 22 | 101 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in flower development. {ECO:0000250|UniProtKB:Q0HA25}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00030 | SELEX | Transfer from AT2G03710 | Download |
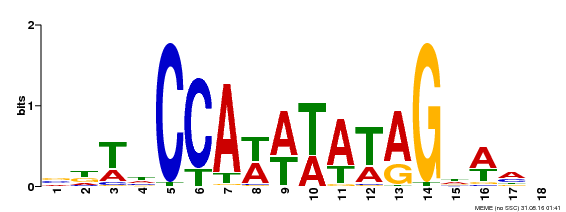
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | evm.model.supercontig_660.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KP942778 | 4e-60 | KP942778.1 Hibiscus sabdariffa var. sabdariffa clone HsMADS13RF1 MADS-box 1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021903284.1 | 0.0 | MADS-box protein CMB1 | ||||
| Swissprot | Q8LLR2 | 1e-114 | MADS2_VITVI; Agamous-like MADS-box protein MADS2 | ||||
| TrEMBL | D7T9Z7 | 1e-124 | D7T9Z7_VITVI; Uncharacterized protein | ||||
| STRING | evm.model.supercontig_660.1 | 0.0 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7135 | 24 | 40 | Representative plant | OGRP16 | 17 | 761 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03710.2 | 1e-106 | MIKC_MADS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm.model.supercontig_660.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



