 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm.model.supercontig_929.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Caricaceae; Carica
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 856aa MW: 97287.2 Da PI: 7.8868 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 161.5 | 1.5e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlyc 83
+e ++rwl+++ei+a+L n++ +++ ++ + pksg+++L++rk++r+frkDG++wkkk dgktv+E+he+LKvg+ e +++
evm.model.supercontig_929.1 30 EEaRTRWLRPNEIHAVLCNYKYFSVNVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKNDGKTVKEAHEHLKVGNEERIHV 111
5559****************************************************************************** PP
CG-1 84 yYahseenptfqrrcywlLeeelekivlvhylevk 118
yYah+++nptf rrcywlL+++le+ivlvhy+e++
evm.model.supercontig_929.1 112 YYAHGQDNPTFVRRCYWLLDKKLEHIVLVHYRETQ 146
********************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 76.482 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.2E-74 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.8E-45 | 31 | 144 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:1.25.40.20 | 1.8E-17 | 488 | 600 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.49E-18 | 489 | 601 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 2.7E-7 | 489 | 567 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.42E-15 | 489 | 597 | No hit | No description |
| PROSITE profile | PS50297 | 16.786 | 505 | 609 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 6.0E-6 | 538 | 567 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.968 | 538 | 570 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.66E-6 | 677 | 752 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.291 | 686 | 712 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 13 | 701 | 723 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.309 | 702 | 731 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0024 | 724 | 746 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.274 | 725 | 749 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 7.6E-4 | 726 | 746 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 14 | 803 | 825 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.766 | 805 | 833 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 856 aa Download sequence Send to blast |
MDGAVSGRLV GSEIHGFRTL DDLDVVTMWE EARTRWLRPN EIHAVLCNYK YFSVNVKPVN 60 LPKSGTIVLF DRKMLRNFRK DGHNWKKKND GKTVKEAHEH LKVGNEERIH VYYAHGQDNP 120 TFVRRCYWLL DKKLEHIVLV HYRETQEVSP ATSGNSNSSS TSDPSTPWFL TEETNCGVDN 180 AYYAQEKESL AESGDRLTLK NHELRLHEIN TLEWDELLAK NDVNSVLPKA GFVQQGQIAV 240 NGPTSDERQL PAYNLSPEKS SFSNSLVPTV QSDILHFDPP VITSASMDVL VNDGLQSQDS 300 FGRWMDYIID DSPGSMSDPT LESPISSVHD SVPSPVISDL LSSVPEQVFT IKDVSPAWAY 360 SNEKTKVFHE LFLSYLNAAS SSELMLLLII MSGCRAGLMT EELFKWEKFQ LQTRLSYLLF 420 STNKSLDILS SKVSPSSLKE ARKFAQKTSH LSNSWAYLMK SIQDNQTSFP EAKDMLFELT 480 LKNRLKEWLI ERILEGCKAT EYDAHGQGVI HLCSILGYTW AIHLFSWSGI SLNFRDKHGW 540 TALHWAAFYG REKMVAVLLS AGAKPNLVTD PTSKNPGGST AADLASKNGY DGLAAFLSEK 600 ALVAQFKDMS LAGNVSGSLQ SSTTDKTNTE NLTEEELYLK DTLAAYRTVA EAASRIQVAF 660 REHSLKVRTK AVEASSPEDD ARNIIAAMKI QHAFRNYETR KRLAAAARIQ HRFRTWKMRR 720 EFLTMRRQAI KIQAAFRGFQ VRRQYRKILW SVGVVEKAIL RWRLKRRGFR GLSVDKCEDV 780 ADQGQESDVE DFYKTSQKQA EERVERSVVS VQAMFRSKKA QEEYRRMKLA HDQAKLEYEC 840 LEDYDTDLLK PQEFR* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
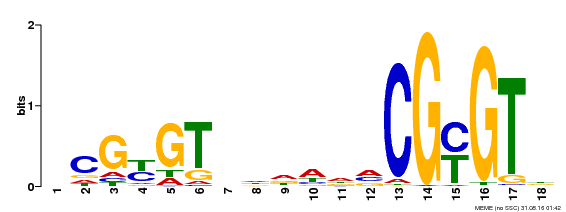
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | evm.model.supercontig_929.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC208139 | 3e-36 | AC208139.1 Populus trichocarpa clone JGIACSB13-D20, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021898630.1 | 0.0 | calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A1R3GK40 | 0.0 | A0A1R3GK40_9ROSI; IQ motif, EF-hand binding site | ||||
| STRING | evm.model.supercontig_929.1 | 0.0 | (Carica papaya) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 | Representative plant | OGRP7351 | 11 | 16 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm.model.supercontig_929.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



