 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00008.135 | ||||||||
| Common Name | AMTR_s00008p00214750, LOC18423305 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 339aa MW: 37250 Da PI: 6.0545 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 60.3 | 3e-19 | 53 | 106 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ +q+++Le+ Fe ++++ e++++LA++lgL+ rqV vWFqNrRa++k
evm_27.model.AmTr_v1.0_scaffold00008.135 53 KKRRLSVDQVKALEKNFEVENKLEPERKAKLAQELGLQPRQVAVWFQNRRARWK 106
566899***********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 127.8 | 4.5e-41 | 52 | 143 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkra 69
ekkrrls +qvk+LE++Fe e+kLeperK++la+eLglqprqvavWFqnrRAR+ktkqlEkdy+ Lk++
evm_27.model.AmTr_v1.0_scaffold00008.135 52 EKKRRLSVDQVKALEKNFEVENKLEPERKAKLAQELGLQPRQVAVWFQNRRARWKTKQLEKDYNLLKAN 120
69******************************************************************* PP
HD-ZIP_I/II 70 ydalkeenerLekeveeLreelk 92
yd+lk + ++++k++e+L +el+
evm_27.model.AmTr_v1.0_scaffold00008.135 121 YDNLKINFDNVKKDKETLMAELR 143
******************99986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.05E-19 | 44 | 110 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.88 | 48 | 108 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.2E-18 | 51 | 112 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.9E-16 | 53 | 106 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.72E-17 | 53 | 109 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.1E-20 | 55 | 115 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 4.1E-6 | 79 | 88 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 83 | 106 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 4.1E-6 | 88 | 104 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 3.6E-16 | 108 | 149 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 339 aa Download sequence Send to blast |
MKRMESSDSL GALISMCPSA EEKNGGGGYS SRGFQSILDG FEEEEECGQA SEKKRRLSVD 60 QVKALEKNFE VENKLEPERK AKLAQELGLQ PRQVAVWFQN RRARWKTKQL EKDYNLLKAN 120 YDNLKINFDN VKKDKETLMA ELRGVKEKVG GRKSGGGGGS GGGDANKEAF PDPQDALAPI 180 PTSKDTEDLK GHTMDGQDRS TGICPDLKDG SDSDSSAILN DENSPLHHPQ SHHYHRPQTL 240 LSSVSSSSAI IAEAAPPTSP PPPLPFLSLH HHHSNNYHIF SSSSSSSSST NGGVTRFYPS 300 PFARMEEHNF LNADESSCNF FADEQAPSLP WYCSDHWN* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 100 | 108 | RRARWKTKQ |
| 2 | 149 | 157 | GGRKSGGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00271 | DAP | Transfer from AT2G22430 | Download |
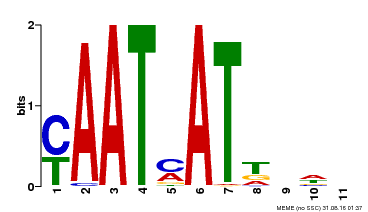
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006827970.1 | 0.0 | homeobox-leucine zipper protein ATHB-6 | ||||
| TrEMBL | W1NIU7 | 0.0 | W1NIU7_AMBTC; Uncharacterized protein | ||||
| STRING | ERM95386 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP129 | 16 | 189 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22430.1 | 1e-57 | homeobox protein 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00008.135 |
| Entrez Gene | 18423305 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



