 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00019.427 | ||||||||
| Common Name | AMTR_s00019p00253580, LOC18435678 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 349aa MW: 37982.7 Da PI: 6.2604 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 52.9 | 5.1e-17 | 260 | 296 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cgt++ Tp++Rrgp g+++LCnaCGl+++ kg+
evm_27.model.AmTr_v1.0_scaffold00019.427 260 CRHCGTSEksTPMMRRGPAGPRSLCNACGLMWANKGT 296
**********************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00979 | 2.7E-8 | 119 | 154 | IPR010399 | Tify domain |
| PROSITE profile | PS51320 | 13.829 | 119 | 154 | IPR010399 | Tify domain |
| Pfam | PF06200 | 5.6E-10 | 121 | 153 | IPR010399 | Tify domain |
| PROSITE profile | PS51017 | 13.285 | 188 | 230 | IPR010402 | CCT domain |
| Pfam | PF06203 | 3.7E-16 | 188 | 230 | IPR010402 | CCT domain |
| PROSITE profile | PS50114 | 9.117 | 254 | 312 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 4.2E-10 | 254 | 307 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 6.18E-12 | 256 | 306 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 2.1E-15 | 259 | 301 | IPR013088 | Zinc finger, NHR/GATA-type |
| CDD | cd00202 | 3.57E-15 | 259 | 303 | No hit | No description |
| PROSITE pattern | PS00344 | 0 | 260 | 287 | IPR000679 | Zinc finger, GATA-type |
| Pfam | PF00320 | 4.1E-15 | 260 | 296 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 349 aa Download sequence Send to blast |
MSDNHSVEIN SQANRNLHQH HTTFSHAYSA AQNPMTDPQV QMHGMHLPGQ IDDVNPEAQI 60 HGGDPMHVHY MQEHGHPLQN NDSGLEDDGE EGGGSEGMEG DGSSDAGNLG DPHTAIPLRS 120 QGTNQLQLSF QGEVYVFDSV APEKVQAVLL LLGGREAPSG LPAVPLYGHQ LSKGFTDPVT 180 QRMNQSHRMQ SLSRFREKRK ERNYDKKIRY SVRKEVAERM QRNKGQFTSS KNVSEDAPPP 240 ELTWESNPTW AAETNEPAIC RHCGTSEKST PMMRRGPAGP RSLCNACGLM WANKGTLRDL 300 SKGGTSLSIQ GHSTNSGEQN EANGTDIKGD SQVTQVVMAL PSNDNSSS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250, ECO:0000269|PubMed:14966217}. | |||||
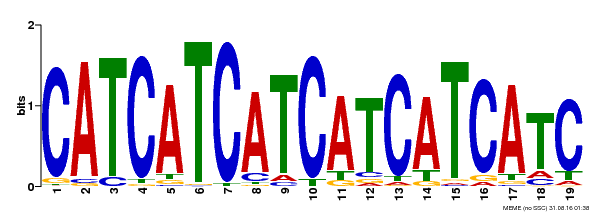
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00198 | DAP | Transfer from AT1G51600 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006845784.1 | 0.0 | GATA transcription factor 24 | ||||
| Refseq | XP_020523606.1 | 0.0 | GATA transcription factor 24 | ||||
| Swissprot | Q8H1G0 | 2e-85 | GAT28_ARATH; GATA transcription factor 28 | ||||
| TrEMBL | W1PIA1 | 0.0 | W1PIA1_AMBTC; Uncharacterized protein | ||||
| STRING | ERN07459 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP831 | 16 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G21175.2 | 4e-79 | ZIM-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00019.427 |
| Entrez Gene | 18435678 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



