 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00021.111 | ||||||||
| Common Name | AMTR_s00021p00158020, LOC18442220 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 376aa MW: 41164.7 Da PI: 9.4427 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 121.3 | 1.2e-37 | 25 | 134 | 2 | 115 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssa 70
+g+kdrhsk++T++g+RdRRvRlsa++a++f+d+qd+LG+d++sk+++WL+++akpai+el+++++ ++
evm_27.model.AmTr_v1.0_scaffold00021.111 25 TGRKDRHSKVCTAKGPRDRRVRLSAHTAIQFYDVQDRLGYDRPSKAVDWLIKKAKPAIDELAELPAWQP 93
79**************************************************************55555 PP
TCP 71 seceaesssssasnsssgkaaksaakskksqksaasalnlakesr 115
+++ + ++ +n++ ++ + +++ ++s+++++ ++nl+++++
evm_27.model.AmTr_v1.0_scaffold00021.111 94 TAA----NLQNPHNKEVAEPSVTKTRARSSSSPKTITQNLSHSHS 134
222....12222222222333333333333333333444444332 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 2.3E-33 | 25 | 135 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 34.543 | 27 | 85 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 376 aa Download sequence Send to blast |
MGERRGGNTV GEIVEVEGGH IVRSTGRKDR HSKVCTAKGP RDRRVRLSAH TAIQFYDVQD 60 RLGYDRPSKA VDWLIKKAKP AIDELAELPA WQPTAANLQN PHNKEVAEPS VTKTRARSSS 120 SPKTITQNLS HSHSTSIAAQ SNPPSSFLLP PMETETMKPF SSSASSSSVG FLNQVQNYPP 180 DLLSRSNSQE LRLSLQSLHD PHHQQMHPIL IQPPAQNPAL QAGIFRFSNE NPLPIHGEQG 240 LFWSEQHHPE FGAARFQRVV AWNSDPSSSS SSSIGFVNSR DVFNSNQSPI IPSFGHHSSF 300 TKQRGPLQSS HYLPIRAREL PPVNNQMASD MPMVSSGFSS GGFSGFRIPA RIQGEEEEHD 360 GVTETSAAAS TSRHP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 2e-20 | 32 | 85 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 2e-20 | 32 | 85 | 1 | 54 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor playing a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164) (PubMed:12931144, PubMed:17307931). Required during early steps of embryogenesis (PubMed:15634699). Participates in ovule develpment (PubMed:25378179). Activates LOX2 expression by binding to the 5'-GGACCA-3' motif found in its promoter (PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:15634699, ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:18816164, ECO:0000269|PubMed:25378179}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00009 | PBM | Transfer from 929286 | Download |
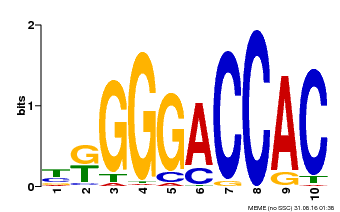
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the miRNA miR-JAW/miR319 (PubMed:12931144, PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:18816164}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011626273.1 | 0.0 | transcription factor TCP4 | ||||
| Swissprot | Q8LPR5 | 6e-72 | TCP4_ARATH; Transcription factor TCP4 | ||||
| TrEMBL | W1PVB3 | 0.0 | W1PVB3_AMBTC; Uncharacterized protein | ||||
| STRING | ERN13972 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP180 | 15 | 163 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G15030.3 | 2e-66 | TCP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00021.111 |
| Entrez Gene | 18442220 |



