 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00022.260 | ||||||||
| Common Name | AMTR_s00022p00216890, LOC18439861 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 622aa MW: 69268.6 Da PI: 5.5454 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 429.4 | 3.5e-131 | 40 | 418 | 1 | 352 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcna 69
+el+krmwkd+++l+r+ker+k ++ +++ k++++++qarrkkmsraQDgiLkYMlk mevc+a
evm_27.model.AmTr_v1.0_scaffold00022.260 40 DELEKRMWKDRIKLQRIKERQKAAA--QQ--AEKPKQKQTSDQARRKKMSRAQDGILKYMLKLMEVCKA 104
79********************854..33..58899********************************* PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STT CS
EIN3 70 qGfvYgiipekgkpvegasdsLraWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslse 138
+GfvYgiipekgkpv+gasd++raWWkekv+fd+ngpaai+ky+a+n +++++ ++ +sl++
evm_27.model.AmTr_v1.0_scaffold00022.260 105 RGFVYGIIPEKGKPVSGASDNIRAWWKEKVKFDKNGPAAIAKYEAENFSAGKAD-----NKGGFNSLQD 168
*********************************************999888555.....45899***** PP
S-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-----GGG--HHHHH CS
EIN3 139 lqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppykkphdlkkawkv 207
lqDTtlgSLLs+lmqhcdppqr+fplekgv+pPWWP G+e+ww+elglsk +ppy+kphdlkk+wkv
evm_27.model.AmTr_v1.0_scaffold00022.260 169 LQDTTLGSLLSSLMQHCDPPQRKFPLEKGVAPPWWPLGNEEWWSELGLSKRLDPPPYRKPHDLKKVWKV 237
********************************************************************* PP
HHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX..XX........X CS
EIN3 208 svLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah..ss........s 266
+vLtavikhmsp+i++i++++rqsk+lqdkm+akes ++l vln+ee ++++s + ss
evm_27.model.AmTr_v1.0_scaffold00022.260 238 GVLTAVIKHMSPNIAKIKKHVRQSKCLQDKMTAKESSIWLGVLNKEEGTIHQLSNEngSSgvsetgtpP 306
*******************************************************96633677677663 PP
XXXXXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX.............................XXXX CS
EIN3 267 lrkqspkvtlsceqkedvegkkeskikhvqavktta.............................gfpv 306
+++++ + +++++dv+g +++ + +++++ +a p
evm_27.model.AmTr_v1.0_scaffold00022.260 307 AFSERKEAEACSNSEYDVDGYEDEPCSVSSKDNGKAskddretclekgdsrenekrlhqefhakePSPQ 375
3346777777778899***77777778888888777888888888888888888888888888776666 PP
XXXXXXXXXXXXXXXXX......XXXXXXX.XXXXXXXXXXXXXXX CS
EIN3 307 vrkrkkkpsesakvsskevsrtcqssqfrgsetelifadknsisqn 352
+rk+++++s+ v+++e ++sq ++++++d+n ++ +
evm_27.model.AmTr_v1.0_scaffold00022.260 376 EARRKRPRTSSTPVNQHEP-LPLEESQDG--VPRNVIPDMNLMEAQ 418
6777777788888777754.444444433..577777777777665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.6E-126 | 40 | 284 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 2.6E-69 | 164 | 292 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 9.94E-59 | 165 | 287 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 622 aa Download sequence Send to blast |
MDQLAMIANE FGDGSDMEVD DIRVDNLAEN EVSDEEIEPD ELEKRMWKDR IKLQRIKERQ 60 KAAAQQAEKP KQKQTSDQAR RKKMSRAQDG ILKYMLKLME VCKARGFVYG IIPEKGKPVS 120 GASDNIRAWW KEKVKFDKNG PAAIAKYEAE NFSAGKADNK GGFNSLQDLQ DTTLGSLLSS 180 LMQHCDPPQR KFPLEKGVAP PWWPLGNEEW WSELGLSKRL DPPPYRKPHD LKKVWKVGVL 240 TAVIKHMSPN IAKIKKHVRQ SKCLQDKMTA KESSIWLGVL NKEEGTIHQL SNENGSSGVS 300 ETGTPPAFSE RKEAEACSNS EYDVDGYEDE PCSVSSKDNG KASKDDRETC LEKGDSRENE 360 KRLHQEFHAK EPSPQEARRK RPRTSSTPVN QHEPLPLEES QDGVPRNVIP DMNLMEAQPI 420 SVQPPPQNIQ LGSAPLPLED PTQLPHQLQD SQFHNVPGEF SGMASANIVA RSMSVKGQPL 480 LFAGAPNTSL HTKATFPAYD SSPGRYGFSH AQSQMQMVNG GHDLRQDGGE IRDENQVFGH 540 VLTLEGRTHE IVGDVNPLVK DAFHSEPDKL VESRFGSPMH DLSLVYGFHS PFDFGIGDTG 600 SLEAADLDFS LEDDLIQYFG A* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 7e-66 | 166 | 295 | 12 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
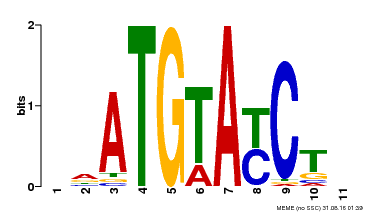
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006850080.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-158 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | W1PVF3 | 0.0 | W1PVF3_AMBTC; Uncharacterized protein | ||||
| STRING | ERN11661 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP563 | 16 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-151 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00022.260 |
| Entrez Gene | 18439861 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



