 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00026.137 | ||||||||
| Common Name | AMTR_s00026p00244220, LOC18438635 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 399aa MW: 45116.2 Da PI: 5.9195 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 29 | 1.8e-09 | 326 | 360 | 21 | 55 |
HSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 21 knrypsaeereeLAkklgLterqVkvWFqNrRake 55
k +yps++e+ LA+++gL+++q+ +WF N+R ++
evm_27.model.AmTr_v1.0_scaffold00026.137 326 KWPYPSETEKVALAESTGLDQKQINNWFINQRKRH 360
569*****************************885 PP
| |||||||
| 2 | ELK | 31.4 | 4.1e-11 | 280 | 301 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELK++Ll+KYsgyLg+Lk E+s
evm_27.model.AmTr_v1.0_scaffold00026.137 280 ELKNHLLKKYSGYLGNLKLELS 301
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 4.4E-21 | 136 | 180 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 5.3E-23 | 137 | 178 | IPR005540 | KNOX1 |
| SMART | SM01256 | 2.7E-29 | 187 | 238 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 3.3E-24 | 192 | 236 | IPR005541 | KNOX2 |
| PROSITE profile | PS51213 | 10.72 | 280 | 300 | IPR005539 | ELK domain |
| Pfam | PF03789 | 1.6E-8 | 280 | 301 | IPR005539 | ELK domain |
| SMART | SM01188 | 1.1E-5 | 280 | 301 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.779 | 300 | 363 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.41E-19 | 301 | 375 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 9.5E-13 | 302 | 367 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 6.3E-28 | 305 | 365 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.03E-11 | 312 | 364 | No hit | No description |
| Pfam | PF05920 | 2.7E-16 | 320 | 359 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 338 | 361 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001708 | Biological Process | cell fate specification | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 399 aa Download sequence Send to blast |
MEHHQLNNEN RRGAFIYTTS PFYGGENMEC HGPDTTTTSM MALMTQNQDQ QPQLHQRLMA 60 PLPIHLLRGH ESQSPQSFHL SSQSLKAEPA GFDRVVRSEH YRAHPGEVVA PRVYAMMLQS 120 HEVENGSNNN TNNDDNAIKA KIVAHPQYSS LLEAYMDCQR VGAPPEVVDR LSAISRELET 180 RQHSSASVGA AGSDPELDQF MEAYYDMLVK YREELTRPIQ EATEFLRRIE SELNSISNGS 240 VRLFSSDDKG DGADSSEEDP DGSGGETELP EIDPRAEDRE LKNHLLKKYS GYLGNLKLEL 300 SKKKKKGKLP KDARQTLLSW WDLHYKWPYP SETEKVALAE STGLDQKQIN NWFINQRKRH 360 WKPSEDMPYV VMDAHHSQNA ALYIEGHLVG DGSYRLGP* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably binds to the DNA sequence 5'-TGAC-3'. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
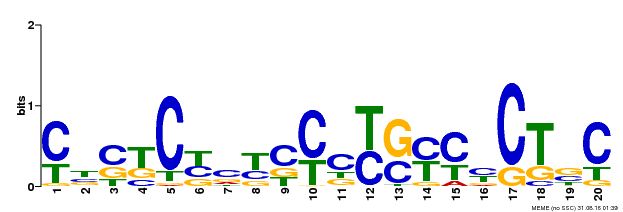
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006848881.1 | 0.0 | homeotic protein knotted-1 isoform X2 | ||||
| Swissprot | O04135 | 1e-137 | KNAP2_MALDO; Homeobox protein knotted-1-like 2 | ||||
| TrEMBL | W1PKD2 | 0.0 | W1PKD2_AMBTC; Uncharacterized protein | ||||
| STRING | ERN10462 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP167 | 17 | 148 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 1e-120 | KNOTTED-like from Arabidopsis thaliana | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00026.137 |
| Entrez Gene | 18438635 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



