 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00045.145 | ||||||||
| Common Name | AMTR_s00045p00141060 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 782aa MW: 83802.9 Da PI: 7.133 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.5 | 3.6e-16 | 527 | 573 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
evm_27.model.AmTr_v1.0_scaffold00045.145 527 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 573
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.4E-20 | 520 | 583 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 523 | 572 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.00E-17 | 526 | 577 | No hit | No description |
| SuperFamily | SSF47459 | 2.22E-20 | 526 | 596 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.2E-13 | 527 | 573 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 529 | 578 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 782 aa Download sequence Send to blast |
MTKEKWDFAQ PKMTNCSTDL AFVPDHEFVE LLWENGQIVL QGQTSRNRKD ASSSGYSSHP 60 PKTHLNHNSG DAVLPVASKG GRYGTLESVL HDFSSGQASG CPLVQEMKWP DHEFVELLWE 120 NGQIVLQGQT SRNRKDASSS GYSSHPPKTH LNHNSGDAVL PVASKGGRYG TLESVLHDFS 180 SGQASGCPLV QEDEMVPWLN YPLDDSLERD YCSEFFPELS GVNLTPLSKP TNVVVSEKAC 240 NLDTTSHKNL SMEHSSQAPS RLRALQPFNS QQGQPSNPIL RQFPSNTHKS SCGNSSSPGA 300 NPAIGIVNSK TEQPNHGSTR PPQPTHTNLM NFSHFSRPAA LVKANLQSMG MAGRLKNNND 360 KASVTDSNPV ESSIVDSAVV SKSGKTKLDQ QCQASLHPTK AKDISSSPAR EKQSVEEDVM 420 YVEDASTSQN RSPDRTLCPS SSFAASTALG ISESVKAVEP VVASSSVCSG NSGGRVSKEP 480 RHGLKRKVGE QEESGYQSED VEGESVGTRK QATGRSATTK RSRAAEVHNL SERRRRDRIN 540 EKMRALQELI PNCNKVDKAS MLDEAIEYLK TLQLQVQIMS MASGMCMPPM MMTAGMQHMP 600 PPHMTHFSPM GVGMGMGMGI GMGMGMGMLD IGGSPGCPLI PLPSIRGPQI PCSSISGPMG 660 LPGMPGSNLQ MYGMPMARAP FLPFSGFPQA KAVAGPDISA PTSSGAFSEL NPPSSSKDQM 720 QNLISPNVQQ KVTDSQQLEA SNQVISEHSA QPNLVPGHNQ SSQVIGSNRN GVAPNGTTSY 780 D* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 531 | 536 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
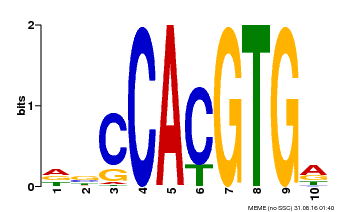
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020520521.1 | 0.0 | transcription factor PIF3 | ||||
| TrEMBL | W1P3A7 | 0.0 | W1P3A7_AMBTC; Uncharacterized protein | ||||
| STRING | ERN02066 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP258 | 16 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-41 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00045.145 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



