 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00049.175 | ||||||||
| Common Name | AMTR_s00049p00181100, LOC18441992 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1317aa MW: 146343 Da PI: 7.2612 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13.4 | 0.00023 | 1287 | 1313 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
evm_27.model.AmTr_v1.0_scaffold00049.175 1287 YVCKdnGCGRTFRFVSDFSRHKRKtgH 1313
99********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 3.1E-18 | 15 | 56 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.409 | 16 | 57 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 2.4E-14 | 17 | 50 | IPR003349 | JmjN domain |
| SuperFamily | SSF51197 | 3.3E-26 | 105 | 143 | No hit | No description |
| SMART | SM00558 | 1.4E-47 | 173 | 342 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 34.029 | 173 | 342 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 3.3E-26 | 191 | 341 | No hit | No description |
| Pfam | PF02373 | 1.1E-35 | 206 | 325 | IPR003347 | JmjC domain |
| SMART | SM00355 | 11 | 1204 | 1226 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 5.4E-6 | 1226 | 1263 | No hit | No description |
| SMART | SM00355 | 0.046 | 1227 | 1251 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.57 | 1227 | 1256 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-4 | 1228 | 1255 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1229 | 1251 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-8 | 1256 | 1281 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.177 | 1257 | 1286 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0014 | 1257 | 1281 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1259 | 1281 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 8.97E-10 | 1267 | 1309 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.4E-9 | 1282 | 1310 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.23 | 1287 | 1313 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.593 | 1287 | 1316 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1289 | 1313 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1317 aa Download sequence Send to blast |
MTDTEVLPWL KSLSLAPEYR PTMAEFQDPI AYIYKIEKEA SSYGICKIIP PLPPSPKKTA 60 IANLNHSLSA LSPSCTFSTR QQQIGFCPRK PRPSATVVQK PVWQSGETYT LEQFEAKAKA 120 FAKSRLKNPN SRPLAIETQF WKATADKPIT VEYANDMPGS AFGDPVEQGV RRVIGESEWN 180 MKVVSRAKGS VLRFMPEEIP GVTSPMVYVA MLFSWFAWHV EDHDLHSLNY LHLGSSKTWY 240 GVPRDAAFAF EEVIRAHGYG GQVNPLVAFA MLGEKTTVMS PQVLIDAGVP CCRLVQNAGE 300 FVVTFPRAYH SGFSHGFNCG EAANIATPEW LRVAKDAAIR RASINYPPMV SHIQLLHALA 360 LSFHSRIPSS ITVEPWSSRL KDKMKGEGEI VVKDLFLQNM IETNDLLHTL SEKGSMCLLL 420 PPSILCETSH VKSKTGLSSN NSEEMSESSN LALRHMTGFD PRKGKAYPLI EGKRVTSIKA 480 RFHKGNHGSY APINRNECLN DSTLSCDLQL DQGLLSCVTC GVLGFACMAV IQPSEAAARN 540 LQSGNCSFLS DQCGGSGLTS DAYPTAEGNA NDSDLNSCSG YTKTDERDDQ DDSQIYSTSH 600 QDCNPPLIKD EKDGQNAYPR TLEKSDPSTA DHNISSLALL ASAYGNASDS EEDEAIQHDI 660 TMHTNEVSPI DTSIACIGTQ QSMPVCAYLP PILRSKDELQ CGDSVLLCSS SPHQNEAANI 720 INHPITNSLS GNEVAAQTSS SFHITNKFSE NSSAKNNNNI PLSRTPNVYQ RRTELVHPDC 780 MPSNCNVPFE QLGCVSNGGP TENVNETAIV NCIDTVRNNR NNSEWQDIDK DSSRMHIFCL 840 EHAMEAEKQL QLMGGANILL LCHSDYPKIE EKAKSIAEEM GVIHSWNGIV FKEASLEDLE 900 RLRVVLEEED DETSHGNGDW AVKLGVNLYH TSNLSRSPLY SKQMPYNSVL YEVLGCNSPD 960 DSSPPGPRPR GRYARQKKIV VAGKWCGKVW MVNQVHPYLS NGKHLKEQHV VLTTKELEND 1020 SKPGERNLDP NQAQKSREDV SEPDEAGTSR DSDPEAVKAS GSLERVSSLD ASLKRKRFTK 1080 RRMSLRRVCS KRPKFCEWGA AGMEGIDPAS EEHDEIGKEP REGETPRSAM KKWNQLDVSP 1140 KGEADEGGPS TRLRQRPRKP QPTSNDETEV PYKRCVRKKR EKKIPESGNK EAREIKAPGD 1200 EDAYQCDIDG CSMGFGTKQE LVIHKRNQCA VKGCGKKFFS HKYLLQHRRV HLDDRPLKCP 1260 WKGCKMTFKW AWARTEHIRV HTGDRPYVCK DNGCGRTFRF VSDFSRHKRK TGHFGK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 1e-81 | 2 | 365 | 1 | 355 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 1e-81 | 2 | 365 | 1 | 355 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 1075 | 1082 | KRFTKRRM |
| 2 | 1176 | 1181 | RKKREK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
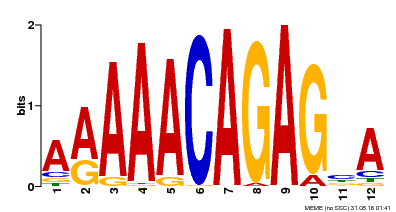
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006852278.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Refseq | XP_020527617.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | W1Q0V7 | 0.0 | W1Q0V7_AMBTC; Uncharacterized protein | ||||
| STRING | ERN13745 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP4401 | 12 | 17 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00049.175 |
| Entrez Gene | 18441992 |



