 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00057.23 | ||||||||
| Common Name | AMTR_s00057p00042800, LOC18445083 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 910aa MW: 102683 Da PI: 6.8662 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 157.5 | 2.6e-49 | 41 | 155 | 4 | 117 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhek 72
e ++rwl+++ei+aiL n++ ++++ ++ + p++g+++L++rk++r+frkDG+ wkkk+dgktvrE+he+
evm_27.model.AmTr_v1.0_scaffold00057.23 41 EaTTRWLRPNEIHAILSNYKLFNVHVKPLDSPENGTIVLFDRKMLRNFRKDGHVWKKKQDGKTVREAHEQ 110
5489****************************************************************** PP
CG-1 73 LKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylev 117
LKvg+ e +++yYah+ ++p+f rr+ywlL++e+ +ivlvhy+++
evm_27.model.AmTr_v1.0_scaffold00057.23 111 LKVGNAERIHVYYAHGLDKPKFARRSYWLLDSEHAHIVLVHYRDT 155
******************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 65.993 | 35 | 161 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.8E-64 | 38 | 156 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.3E-44 | 41 | 154 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 5.0E-4 | 359 | 446 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.48E-13 | 360 | 446 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 2.4E-4 | 360 | 445 | IPR002909 | IPT domain |
| Gene3D | G3DSA:1.25.40.20 | 4.1E-15 | 548 | 661 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.75E-14 | 548 | 658 | No hit | No description |
| SuperFamily | SSF48403 | 3.51E-15 | 557 | 661 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.831 | 566 | 670 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.327 | 599 | 631 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0013 | 599 | 628 | IPR002110 | Ankyrin repeat |
| Pfam | PF13637 | 5.2E-5 | 601 | 658 | No hit | No description |
| SMART | SM00248 | 2500 | 638 | 667 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 4.78E-6 | 735 | 812 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 6.65 | 746 | 772 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.17 | 746 | 762 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 4.2 | 761 | 783 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.078 | 762 | 791 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.21 | 784 | 806 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.615 | 785 | 809 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0018 | 787 | 805 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 910 aa Download sequence Send to blast |
MKTQGLSRRD LLEGSEDCLL GSEIHGFRTL EELQIGKIPT EATTRWLRPN EIHAILSNYK 60 LFNVHVKPLD SPENGTIVLF DRKMLRNFRK DGHVWKKKQD GKTVREAHEQ LKVGNAERIH 120 VYYAHGLDKP KFARRSYWLL DSEHAHIVLV HYRDTSEDTA NGTISSRKES KEAFSLLGRP 180 RALHLSPSTP STSSSGSSYL DQPVISSEEN NPGADSTAYK SAEMGNQFLG AGETRNFESI 240 IHEINTYEWD DLLGILNTAP DEGRDLSSER AQCDQKDSVA IVNDVLLSNT EGRHASHLMD 300 SNFKRNCVDQ EAVNQYTQSA PSETLINDDA LQPQDTYNTI MNYVQNNPPG SPGQGQVFYI 360 TDISPAWAFA TEETKVIVTG YLLETQATLE ELNWFCVIGD VTVPAEIIQP GVLRFIAPPH 420 SSGVYDFYLT LDCRTPSSQV YSFEYRPFSS NQVNNIGFSP KVDELEWDDF DVQVRLAHLL 480 FSTTKGLAVL YSKETPKAMR EARRFSSLIS SYEKEWTSLL KSTKSKNISL SQAKSNLMEL 540 TLKGKLWEWL LETVVEGRRP AVKDSHGQGV IHLCAILGYA WAIYPISASG MSINFRDASG 600 WTALHWAAYC GREQMAAVLL STGANASLVS DPTPDCPGGC TAADLALRKG FEGMAAYLGE 660 KALTAHFRDM HISGHLGSSD EASLENTNDT QNLTEEELCI RDSMAAIRTA ANAASQIQAA 720 FREHSFNLRI KAVQLAKPDY EWAIISAMKI QNAYRAHSQR KRTAAAVRIQ HRFRTWKIRR 780 EFLNMRRQVI KIQAIFRGYQ VRRQYCKILW SVGILEKGIL RWRQKRKGLR GLQVEPTTPV 840 EIEEDGVPVV EEDFFRIGRK LAEERVDRSF NSVQVMFQSD QARRDYRRLK LKVDAAMLEH 900 DTVHDTDQD* |
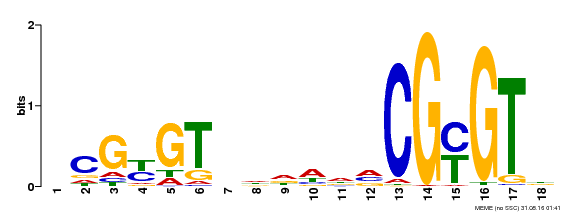
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006855291.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X3 | ||||
| TrEMBL | U5D8P3 | 0.0 | U5D8P3_AMBTC; Uncharacterized protein | ||||
| STRING | ERN16758 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP7351 | 11 | 16 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00057.23 |
| Entrez Gene | 18445083 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



