 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | evm_27.model.AmTr_v1.0_scaffold00099.164 | ||||||||
| Common Name | AMTR_s00099p00163300 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; basal Magnoliophyta; Amborellales; Amborellaceae; Amborella
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 323aa MW: 37022.3 Da PI: 7.1276 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 157.7 | 4.9e-49 | 13 | 140 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkya 69
l+pGfrFhPt+eelv +yL++kvegk++++ e i+ +d+y+++Pw+Lp+++ +ekew+f+++rd+ky+
evm_27.model.AmTr_v1.0_scaffold00099.164 13 LMPGFRFHPTEEELVEFYLRRKVEGKRFSV-ELITFLDLYRYDPWELPAHAVIGEKEWFFYVPRDRKYR 80
689***************************.89***************8777899************** PP
NAM 70 tgkrknratksgyWkatgkdkevlsk.kgelvglkktLvfykgrapkgektdWvmheyrl 128
+g+r+nr+t+sgyWkatg d+ + + +++ +glkktLvfy+g+apkg++++W+m+eyrl
evm_27.model.AmTr_v1.0_scaffold00099.164 81 NGDRPNRVTTSGYWKATGADRMIRGErSSRPIGLKKTLVFYSGKAPKGTRSSWIMNEYRL 140
***********************99857788***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 9.42E-55 | 11 | 169 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 55.408 | 13 | 170 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.2E-25 | 15 | 140 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 323 aa Download sequence Send to blast |
MNDAEKEGGE EVLMPGFRFH PTEEELVEFY LRRKVEGKRF SVELITFLDL YRYDPWELPA 60 HAVIGEKEWF FYVPRDRKYR NGDRPNRVTT SGYWKATGAD RMIRGERSSR PIGLKKTLVF 120 YSGKAPKGTR SSWIMNEYRL PQHETTSADH NHHIFQKQKV EISLCRVYKR AGGEDLSTIP 180 VPRFPTRLGN PSGRTALDEN TRAIDHASDE SLQFGGGDAT PSSSSSSSTR VLHHQYNNLD 240 NSMVTPPCND TLLTLMAPTT TVAEELHRLV EYQQIYVQAN QSQTSQSLLN TFSSSLPNLS 300 DKLWEWNPVP EPSRDYYHFK EQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| 3swm_B | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| 3swm_C | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| 3swm_D | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| 3swp_A | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| 3swp_B | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| 3swp_C | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| 3swp_D | 3e-54 | 13 | 170 | 20 | 168 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that acts as a floral repressor. Controls flowering time by negatively regulating CONSTANS (CO) expression in a GIGANTEA (GI)-independent manner. Regulates the plant cold response by positive regulation of the cold response genes COR15A and KIN1. May coordinate cold response and flowering time. {ECO:0000269|PubMed:17653269}. | |||||
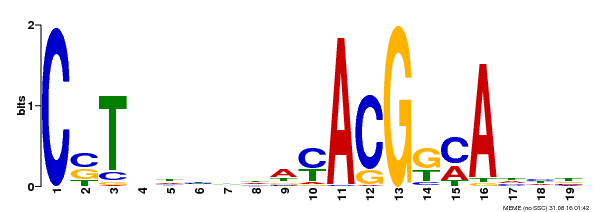
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00257 | DAP | Transfer from AT2G02450 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian regulation with a peak of expression at dawn under continuous light conditions (PubMed:17653269). Circadian regulation with a peak of expression around dusk and lowest expression around dawn under continuous light conditions (at protein level) (PubMed:17653269). {ECO:0000269|PubMed:17653269}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006836956.2 | 0.0 | NAC domain-containing protein 35 | ||||
| Swissprot | Q9ZVP8 | 9e-96 | NAC35_ARATH; NAC domain-containing protein 35 | ||||
| TrEMBL | W1NYP6 | 0.0 | W1NYP6_AMBTC; Uncharacterized protein | ||||
| STRING | ERM99809 | 0.0 | (Amborella trichopoda) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP17 | 15 | 800 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02450.1 | 1e-100 | NAC domain containing protein 35 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | evm_27.model.AmTr_v1.0_scaffold00099.164 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



