 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | gw1.4.455.1 | ||||||||
| Common Name | CHLNCDRAFT_15266 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Chlorellales; Chlorellaceae; Chlorella
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 88aa MW: 10467.9 Da PI: 11.7011 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 35.4 | 2.5e-11 | 2 | 33 | 15 | 47 |
HHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 15 avkqlGggtWktIartmgkgRtlkqcksrwqky 47
+v+ +G +W++Ia+++ gR +kqc++rw++
gw1.4.455.1 2 LVRDHGQSNWSLIAKHFS-GRIGKQCRERWHNQ 33
899***************.************96 PP
| |||||||
| 2 | Myb_DNA-binding | 47.1 | 5.4e-15 | 40 | 82 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
r +W E+ ll++a+k+ G++ W+ Ia+ ++ gRt++ +k++w+
gw1.4.455.1 40 REAWEGGEERLLIEAHKRIGNK-WADIAKLIP-GRTENAVKNHWN 82
5678888***************.*********.***********9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00167 | 1.94E-7 | 1 | 32 | No hit | No description |
| SMART | SM00717 | 0.12 | 1 | 36 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-19 | 1 | 41 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 7.0E-10 | 1 | 34 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 15.642 | 1 | 34 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.21E-25 | 13 | 87 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 24.006 | 35 | 88 | IPR017930 | Myb domain |
| SMART | SM00717 | 7.0E-12 | 39 | 87 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-13 | 40 | 82 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.7E-20 | 42 | 88 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.62E-7 | 47 | 85 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 88 aa Download sequence Send to blast |
RLVRDHGQSN WSLIAKHFSG RIGKQCRERW HNQLRPDIRR EAWEGGEERL LIEAHKRIGN 60 KWADIAKLIP GRTENAVKNH WNATLRRK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 4e-39 | 1 | 88 | 17 | 104 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 4e-39 | 1 | 88 | 17 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 4e-39 | 1 | 88 | 17 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor required for female gametophyte fertility. Acts redundantly with MYB64 to initiate the FG5 transition during female gametophyte development. The FG5 transition represents the switch between free nuclear divisions and cellularization-differentiation in female gametophyte, and occurs during developmental stage FG5. {ECO:0000269|PubMed:24068955}. | |||||
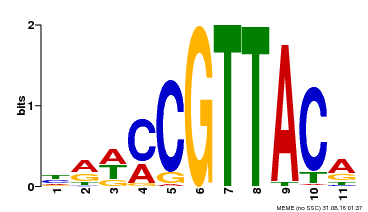
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00567 | DAP | Transfer from AT5G58850 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_005850135.1 | 7e-58 | hypothetical protein CHLNCDRAFT_15266, partial | ||||
| Swissprot | Q9FIM4 | 9e-39 | MY119_ARATH; Transcription factor MYB119 | ||||
| TrEMBL | E1Z842 | 2e-56 | E1Z842_CHLVA; Uncharacterized protein (Fragment) | ||||
| STRING | XP_005850135.1 | 3e-57 | (Chlorella variabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP15 | 16 | 114 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58850.1 | 4e-41 | myb domain protein 119 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 17357714 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



