 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | gw1.5.634.1 | ||||||||
| Common Name | CHLNCDRAFT_17292 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Trebouxiophyceae; Chlorellales; Chlorellaceae; Chlorella
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 268aa MW: 28788.6 Da PI: 8.1903 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.1 | 3.2e-05 | 246 | 268 | 2 | 23 |
EETTTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCpdCgksFsrksnLkrHirt.H 23
+Cp+Cg+ F + +L+ H H
gw1.5.634.1 246 RCPHCGREFADAVQLVAHVEQqH 268
7*****************87655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF118310 | 1.75E-5 | 2 | 34 | No hit | No description |
| Gene3D | G3DSA:4.10.1110.10 | 7.4E-5 | 3 | 33 | IPR000058 | Zinc finger, AN1-type |
| SMART | SM00154 | 0.41 | 5 | 41 | IPR000058 | Zinc finger, AN1-type |
| Pfam | PF01428 | 1.1E-8 | 5 | 33 | IPR000058 | Zinc finger, AN1-type |
| Gene3D | G3DSA:4.10.1110.10 | 1.8E-10 | 69 | 125 | IPR000058 | Zinc finger, AN1-type |
| SuperFamily | SSF118310 | 1.83E-10 | 77 | 127 | No hit | No description |
| PROSITE profile | PS51039 | 8.582 | 84 | 137 | IPR000058 | Zinc finger, AN1-type |
| SMART | SM00154 | 0.014 | 87 | 128 | IPR000058 | Zinc finger, AN1-type |
| Pfam | PF01428 | 5.8E-10 | 87 | 124 | IPR000058 | Zinc finger, AN1-type |
| SMART | SM00355 | 0.17 | 198 | 221 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.681 | 198 | 226 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 200 | 221 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 9.972 | 245 | 268 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.019 | 245 | 268 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 247 | 268 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 268 aa Download sequence Send to blast |
IGRHCAHADC HQLDFLPFRC DRCSKVYCLE HRVCPCSKDG DTVLVCPLCA KAVVCPPGGD 60 VELVYDRHTR TECDPSNYDK VHRKPRCPVG GCKEKLTTIN SYTCRECGVT VCLKHRLAAD 120 HKCQVPRQQP APVAAGGGSS SRRPTVAQKA AAVAAKTKDS IQSQLQQYRQ QRKGGPPGAA 180 GVVDLTGSSP TATPNGAEQC PQCGVRFATV QQLIEHAEAA HSGGWSSGAV NMQQQQHVPP 240 AGGLERCPHC GREFADAVQL VAHVEQQH |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00306 | DAP | Transfer from AT2G41835 | Download |
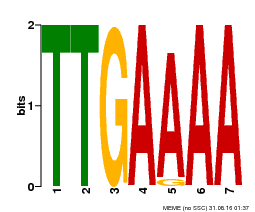
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_005849841.1 | 0.0 | hypothetical protein CHLNCDRAFT_17292, partial | ||||
| TrEMBL | E1Z996 | 0.0 | E1Z996_CHLVA; Uncharacterized protein (Fragment) | ||||
| STRING | XP_005849841.1 | 0.0 | (Chlorella variabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP4812 | 4 | 4 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G41835.1 | 4e-53 | C2H2 family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 17357264 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



