 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | kfl00073_0220 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Klebsormidiophyceae; Klebsormidiales; Klebsormidiaceae; Klebsormidium
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 1116aa MW: 119045 Da PI: 5.6652 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 44.2 | 3.5e-14 | 893 | 978 | 1 | 83 |
EEEE-..-HHHHTT-EE--HHH.HTT...---..--SEEEEEETTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh...ggkkeesktltledesgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFk 83
f+kvlt sd++ +gr++lpk+ ae h ++ke++ +t+ed + ++W++++++ ++ks+ yv++ + +Fv++ gL+ gD +v
kfl00073_0220 893 FEKVLTASDTSPLGRIILPKSQAELHlpkVLDKEGG-LITVEDADRNRWQLRYRFwpNNKSRIYVFE-HTADFVQSRGLRPGDYLVVC 978
99**************************96666555.9***************99655555558888.9***************9976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.0E-5 | 379 | 441 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-6 | 384 | 443 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:2.40.330.10 | 4.6E-22 | 888 | 995 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 3.27E-19 | 890 | 988 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.76E-19 | 891 | 976 | No hit | No description |
| Pfam | PF02362 | 3.0E-11 | 893 | 978 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 9.756 | 893 | 993 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.9E-13 | 893 | 993 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1116 aa Download sequence Send to blast |
MRPEAFEPAR SQGPVDTDAD SRSSSCRHAA ETGPPEVESA SRAGNVNSQG LVGTGVAEQL 60 DQLGDAMLAE VEAALAAEAR GDGFYGANKL GVNEFGVHEP RKLSRLASLD VLSEILSWEA 120 FGGSEKHEER GVVDSKVGVV DSRPRGGGVE SDVDNNEDDA VLASEKGGGD RQGGKDGVRG 180 DESHQVNRGD VTKVPASVES GGGILKSKSI ESEWGIALVK CGENAGGGTE DASSEGAVQE 240 LLKSYDRALD IKEKEERGGG APSTEPSSGL SQLGEMEVDQ ELMTALKSNV QLRDEHGTLE 300 RAGSGEGSGE QTGGWFGRGS GNRVSGNRLQ SRRPHSTPTS PVLSPLPSPK MTLRSQSFKE 360 SVQVGGYSRV PVDVPKPQPA GENQRMRFSP QQQKTLQEFG TKVNWVGPGV ESMEEAHQIC 420 AEMSMDIKQL KKWISNHRPK ELRSSSTPRP GVQPPWAIVP PLANSAFPGV YSEAALRGLS 480 AAHPALVGHP FYPPAPFFGG PQFPQGAWVP PHYAPGGQAM WPWPASGASL PPQASTPGGA 540 VPPPQQRSDA PGEGGGKPVD VKEIGPGGAE LVRGGSLEAN GGKQVPLDME RLANADNLRQ 600 VLGRKLKDFD PGKIQKLVEI CKTYRQKQSS DSPHERHDGP PKKKQLKSPP EVENQLPFPP 660 PLNLDSATPQ DEYQPQWGDT LLAESTPFGG RSDNLVADPT GWNMGDLANA PFANAPFENA 720 TTLQGDGRAK AEKGLPKPGL AYPNPSMHGE LAQAFDQNFL GGQPRDEMSL LPFPPPSNQF 780 LAHSAAGGPG QQTWPEPGFD AGLANGFFYN PAAHPAAFGA FSPLEMGLPA GGVPRPATSS 840 GKPPRTVRAL HMRGGCTAPF PGTDPKEWRV NEAPGLWVRG PVTPQPMRGK FLFEKVLTAS 900 DTSPLGRIIL PKSQAELHLP KVLDKEGGLI TVEDADRNRW QLRYRFWPNN KSRIYVFEHT 960 ADFVQSRGLR PGDYLVVCRA DEGHLILNDR RRPESPEGDP SERRATSEGS RSTVSSGSSC 1020 NRGTPDARTA SEHAESAALT SAAGLTSFRA GPPPLPLQQA GRQLSMEEVT RWAMSGDGHE 1080 MMDCVNLDHG EEPHVLELTQ ELQSLHSSGG LPRLGS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9b_A | 9e-24 | 890 | 1002 | 2 | 116 | B3 domain-containing transcription factor FUS3 |
| 6j9b_D | 9e-24 | 890 | 1002 | 2 | 116 | B3 domain-containing transcription factor FUS3 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
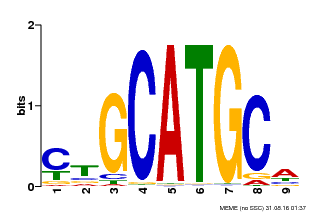
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A1Y1HRB4 | 0.0 | A0A1Y1HRB4_KLENI; Uncharacterized protein | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 1e-24 | B3 family protein | ||||



