 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | kfl00189_0180 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Klebsormidiophyceae; Klebsormidiales; Klebsormidiaceae; Klebsormidium
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 1072aa MW: 114695 Da PI: 9.0234 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 92.7 | 2.7e-29 | 360 | 417 | 3 | 59 |
-SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 3 DgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
Dg+nWrKYGqK kg+++pr+YYrCt+a gCpv+k+v+rsa+dp+v+++tYe Hnh+
kfl00189_0180 360 DGCNWRKYGQKIAKGNPCPRAYYRCTQAaGCPVRKQVQRSADDPSVLVVTYEHMHNHP 417
9**************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 5.8E-29 | 345 | 417 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.79E-26 | 352 | 418 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 27.439 | 353 | 419 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.9E-36 | 358 | 418 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.5E-26 | 360 | 417 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1072 aa Download sequence Send to blast |
MEALLNDESR SQGNNHSPLQ QEGSDNLYSG FIDRFPSFDS MFGPKQRRDS PQSSDAYTQR 60 ENERLSRARI EAEEIERLRK GALSPLKGAR AKMEDEMDAQ IRGGDRRNGA ALTRALNLSR 120 LGAPQQNWAP VKDEFNQRGL MKPAEDNQEK EAENGDMGFV DKEVTTWYSV SNRQAVGAGD 180 YATSAALIMQ QKLWEAMQEI QRLTEEGQAR EKLCAKLLQQ NAFLQAQIKQ YQDWSGLPAA 240 DRTRWKSPEP FAQPTHYKNE PRSPSLAPVD VPPVGNRPGG HQAGGALFER TTLGALSPHR 300 PLSPQCMRAD PLNLAVKPEA ERAVGHIPDM ENLQKVMQEN AEKSAKVALV MKSNAITIFD 360 GCNWRKYGQK IAKGNPCPRA YYRCTQAAGC PVRKQVQRSA DDPSVLVVTY EHMHNHPLSS 420 AAAPMAEATS SLASSYLCNV TSSNPNASAF AGVTASVDEG HASSNYAPNA NIPTITVDTI 480 NQAGSEWALR IAAARASAAL PNGGAKGETL NPQAKGAPSR AGVFSTVESV LKQQVKRAQA 540 AAPKDPAEPL NPRAKSLSGE KAQRTSSGEK KPKKKASSDS KGKAPLTLPV KAELMADAQT 600 PRFVQTLKTP RSAAASTTTT PFPPASPAVD APAGASLLTS LQTITERLQE LPGDEKPSGI 660 LEKLPTRGAK KRSRSRTVSA DTSVPVVPST TDDGARASAH GAAAFEASER GDQAVPEPPR 720 SLQLLRTSSR TSSDNLLHAL LSGAKYTPGP SPQNSCLQEQ GLSTLMGSGL SSFGLFAAAR 780 SGVKETDHDA DEVQSPRRSD GGAPDGDAAA KPQTSRVARP WPSYFQPGGS TVKGDKPTDA 840 SPEQSPLPWA SFHPFGLLSS TYNSYGTNFT PGPFTGGFGA HFSNRGNIFP FGPTPEMRDE 900 KTPGASAREE TAAAGGDVRG RPRSLSLNLG PVSPPMVSKS NGDPARTLEH QFDGDDSPLA 960 LVRRSPENNF TGLLTAVESP ANEELVRERK RARVASPGVE VGDREKVVVL DQEKRPGNAQ 1020 PGPAGTVGGE LPTFNLMGAA ITHGRIEVKT VENARLALEQ FLTAIRDTGL EK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-18 | 360 | 418 | 19 | 76 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-18 | 360 | 418 | 19 | 76 | Probable WRKY transcription factor 4 |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 670 | 675 | KRSRSR |
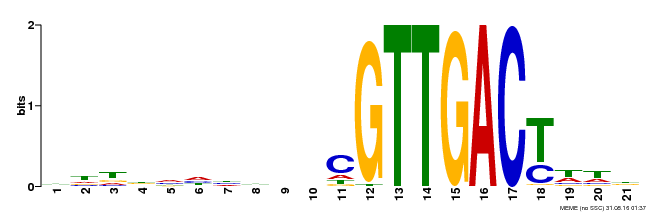
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00428 | DAP | Transfer from AT4G04450 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A1Y1I0L3 | 0.0 | A0A1Y1I0L3_KLENI; WRKY DNA-binding protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Representative plant | OGRP14 | 17 | 875 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G04450.1 | 4e-35 | WRKY family protein | ||||



