 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold01003-augustus-gene-0.45-mRNA-2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 426aa MW: 45764 Da PI: 9.0834 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 72.9 | 4.5e-23 | 281 | 343 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkrerrkq+NRe+ArrsR+RK+ae+eeL+ kv++L+aeN a k+e+e+l+++++klk e+
maker-scaffold01003-augustus-gene-0.45-mRNA-2 281 ERELKRERRKQSNRESARRSRLRKQAETEELAHKVETLTAENGAIKSEIEQLTDNSEKLKLEN 343
89**********************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 6.8E-38 | 1 | 96 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 1.2E-26 | 129 | 260 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 4.3E-18 | 275 | 340 | No hit | No description |
| SMART | SM00338 | 7.4E-22 | 281 | 345 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.0E-21 | 281 | 343 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.024 | 283 | 346 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.69E-11 | 284 | 340 | No hit | No description |
| CDD | cd14702 | 4.41E-20 | 286 | 333 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 288 | 303 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 426 aa Download sequence Send to blast |
MGNNEEGKPT KSEKQSSPAR LDQTNQTNVH VYPDWAAMQA YYGPRVAIPP YYNSTMASGH 60 APHPYMWGTP QPMMPPYGAP YAAIYPHGGL YAHPAVTLGS HSHGQGVPLS PAAVTPMSIE 120 TPTKSGNTDR GLMKKLKEFD GLAMSIGNGT AESLEGGAEH RLSQSAETEG SSDGSDGNTA 180 KANQTRRKRS REETPTTGIG GKTETQASPV TSKEVNTASN KVLGAAVTPA SVTVNLVGNV 240 VSPGMTTLEF RNTPNANTKT TTPSVPQSCT VLPSEGWPQN ERELKRERRK QSNRESARRS 300 RLRKQAETEE LAHKVETLTA ENGAIKSEIE QLTDNSEKLK LENATLMEKL KKAQLGHREE 360 IILNSIGGKR TLPVSTENLL SRVNNSGSIG ISIEEKSDMY DKNSNSGAKL HQLLDASPRA 420 DAVAAG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 297 | 303 | RRSRLRK |
| 2 | 297 | 304 | RRSRLRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the G-box-like motif (5'-ACGTGGC-3') of the chalcone synthase (CHS) gene promoter. G-box and G-box-like motifs are defined in promoters of certain plant genes which are regulated by such diverse stimuli as light-induction or hormone control. | |||||
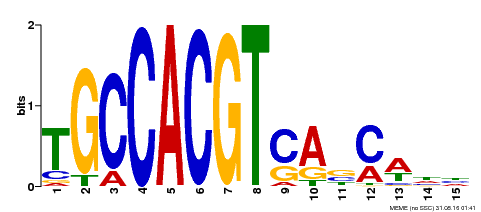
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00318 | DAP | Transfer from AT2G46270 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JF428135 | 1e-149 | JF428135.1 Corylus heterophylla bZIP78 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023875514.1 | 0.0 | common plant regulatory factor 1-like isoform X2 | ||||
| Refseq | XP_023875515.1 | 0.0 | common plant regulatory factor 1-like isoform X2 | ||||
| Swissprot | Q99089 | 1e-144 | CPRF1_PETCR; Common plant regulatory factor 1 | ||||
| TrEMBL | A0A2I4F8E0 | 0.0 | A0A2I4F8E0_JUGRE; common plant regulatory factor 1-like isoform X2 | ||||
| STRING | EMJ19254 | 0.0 | (Prunus persica) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46270.1 | 1e-114 | G-box binding factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



