 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold01338-augustus-gene-0.35-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 327aa MW: 37069 Da PI: 5.7325 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 79.9 | 2.6e-25 | 135 | 195 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnhe 59
dD ++WrKYGqKe+ ++++prsY+rCt + gC+++k+v+r +edp++ +itY g H+++
maker-scaffold01338-augustus-gene-0.35-mRNA-1 135 DDDHAWRKYGQKEILNAKHPRSYFRCTRKydqGCRATKQVQRMEEDPQMYQITYIGIHTCK 195
8**************************998899**************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-24 | 123 | 195 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 19.634 | 129 | 192 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.89E-24 | 131 | 195 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.1E-35 | 134 | 196 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.1E-24 | 135 | 195 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009862 | Biological Process | systemic acquired resistance, salicylic acid mediated signaling pathway | ||||
| GO:0009864 | Biological Process | induced systemic resistance, jasmonic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
METTHPWPEG PSTTRNKVIK ELVEGRDFAT QLKSILHKPL EITRDHASPS PSPHELVSKI 60 LRSFTETLSV LASSCESGEV IHQNLAVSHA DSHCDDPRSD DSGESRKRPT PKDRRGCYKR 120 RKTTQTWTTV SRTTDDDHAW RKYGQKEILN AKHPRSYFRC TRKYDQGCRA TKQVQRMEED 180 PQMYQITYIG IHTCKDTLKA PQIIMEPDPW ETYIVSNPPD SMWGNGKPAD HDQALSSSTT 240 LTIKQESKEE ALSDLTDNLS SLGTDNNNNS YLWTDFKDFD LVESAIMSPK LGSDNGDVVS 300 TMYSCTETAS HGLDFCHFDG DFNFDDI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 7e-25 | 128 | 195 | 2 | 69 | OsWRKY45 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 105 | 121 | RKRPTPKDRRGCYKRRK |
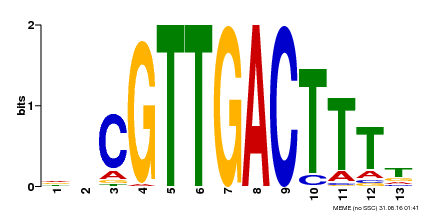
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00408 | DAP | Transfer from AT3G56400 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023894740.1 | 0.0 | probable WRKY transcription factor 70 isoform X1 | ||||
| Refseq | XP_023894741.1 | 0.0 | probable WRKY transcription factor 70 isoform X2 | ||||
| TrEMBL | A0A2N9EI06 | 1e-161 | A0A2N9EI06_FAGSY; Uncharacterized protein | ||||
| STRING | GLYMA04G40130.1 | 1e-93 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4700 | 30 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56400.1 | 8e-46 | WRKY DNA-binding protein 70 | ||||



