 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold02241-augustus-gene-0.25-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 294aa MW: 32165.2 Da PI: 7.5878 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 30.8 | 7e-10 | 7 | 54 | 4 | 47 |
S-HHHHHHHHHHHHHTTTT.....-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 4 WTteEdellvdavkqlGgg.....tWktIartmgkgRtlkqcksrwqky 47
W++eE++ + +a++++ + +W++Ia++++ +t ++k ++q +
maker-scaffold02241-augustus-gene-0.25-mRNA-1 7 WSKEEEKTFENAIAMHWVDedckeQWEKIASMVP-SKTMDELKQHYQLL 54
**********************************.***********965 PP
| |||||||
| 2 | Myb_DNA-binding | 37.2 | 7e-12 | 131 | 175 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT++E+ l++ + ++G+g+W++I+r + Rt+ q+ s+ qky
maker-scaffold02241-augustus-gene-0.25-mRNA-1 131 PWTEDEHRLFLLGLDKFGKGDWRSISRNFVISRTPTQVASHAQKY 175
7*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 9.818 | 1 | 58 | IPR017884 | SANT domain |
| SMART | SM00717 | 4.1E-5 | 3 | 57 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.6E-8 | 7 | 53 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.03E-8 | 7 | 55 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-5 | 7 | 52 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.79E-9 | 7 | 61 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.282 | 124 | 180 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.15E-16 | 126 | 181 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.8E-17 | 128 | 178 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 9.6E-10 | 128 | 178 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-10 | 129 | 174 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.42E-9 | 131 | 176 | No hit | No description |
| Pfam | PF00249 | 2.4E-10 | 131 | 175 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 294 aa Download sequence Send to blast |
MSSGIIWSKE EEKTFENAIA MHWVDEDCKE QWEKIASMVP SKTMDELKQH YQLLVEDVSA 60 IEAGHIPLPN YVGEEASSSN KDCHGFSGAM ASDKRSNCGF GGGFSGLGHD STGHGGKGGS 120 RSEQERRKGI PWTEDEHRLF LLGLDKFGKG DWRSISRNFV ISRTPTQVAS HAQKYFIRLN 180 SMNRDRRRSS IHDITSVNNG DVASHQAPIT GQQTNMNPSS VATIGPPVKH RTQPHMPGLG 240 MYGTSVGHPI AAPPVHMASA VGTPVMLPPG HHPHSHPPYV MPVAYPMAPP PMHQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 2e-14 | 3 | 71 | 7 | 73 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that coordinates abscisic acid (ABA) biosynthesis and signaling-related genes via binding to the specific promoter motif 5'-(A/T)AACCAT-3'. Represses ABA-mediated salt (e.g. NaCl and KCl) stress tolerance. Regulates leaf shape and promotes vegetative growth. {ECO:0000269|PubMed:26243618}. | |||||
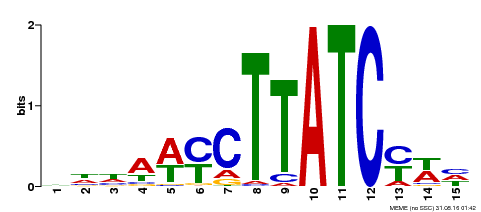
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00191 | DAP | Transfer from AT1G49010 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salicylic acid (SA) and gibberellic acid (GA) (PubMed:16463103). Triggered by dehydration and salt stress (PubMed:26243618). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:26243618}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023913108.1 | 0.0 | transcription factor MYBS1 | ||||
| Swissprot | Q9FNN6 | 4e-82 | SRM1_ARATH; Transcription factor SRM1 | ||||
| TrEMBL | A0A2I4F186 | 0.0 | A0A2I4F186_JUGRE; transcription factor DIVARICATA-like | ||||
| STRING | POPTR_0012s05720.1 | 1e-162 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF10694 | 33 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49010.1 | 1e-79 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



