 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold02381-snap-gene-0.26-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 292aa MW: 32348.3 Da PI: 4.6641 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.6 | 9.2e-13 | 214 | 259 | 6 | 55 |
HHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 6 nerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+++Er RR ri +++ +L+el+P+ +k + a++Le+AveY+k Lq
maker-scaffold02381-snap-gene-0.26-mRNA-1 214 SIAERVRRTRISDRIRKLQELVPNM----DKQTNTADMLEEAVEYVKLLQ 259
689*********************7....688***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.615 | 208 | 258 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.44E-16 | 211 | 274 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 9.35E-11 | 212 | 263 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.8E-16 | 213 | 274 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 7.6E-10 | 214 | 259 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 9.1E-12 | 214 | 264 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 292 aa Download sequence Send to blast |
MQPTPDGRSN SGGGGSGELS GGGGLARFRS APATWLEALL EEEKPVAQKE EREGNVEEEG 60 EEEEEEDEDE EDPLEPTQSL TQLLASSADP SLLVFDSNIF RQNSSPAEFF GNTDSEAYFS 120 NFGLNPNPPN YNNYDSHSNS NTIHGGGGTG TKRSRELNHH FTAKFPSHHH LKGEQSSGGG 180 GVGSFIDMEM EKILEDSVPC RVRAKRGCAT HPRSIAERVR RTRISDRIRK LQELVPNMDK 240 QTNTADMLEE AVEYVKLLQK QIQDLSEQQQ RCKCVVKDFT EESIKDSSHS HV |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00181 | DAP | Transfer from AT1G35460 | Download |
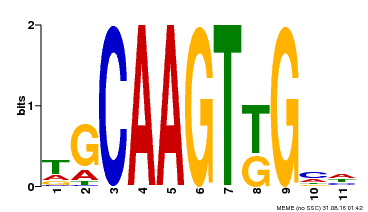
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023889026.1 | 1e-156 | transcription factor bHLH80-like isoform X2 | ||||
| Swissprot | Q9C8P8 | 2e-68 | BH080_ARATH; Transcription factor bHLH80 | ||||
| TrEMBL | A0A2I4HJU9 | 1e-86 | A0A2I4HJU9_JUGRE; transcription factor bHLH80-like isoform X2 | ||||
| STRING | Gorai.013G138300.1 | 2e-79 | (Gossypium raimondii) | ||||
| STRING | XP_004168264.1 | 2e-79 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9132 | 30 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G35460.1 | 3e-52 | bHLH family protein | ||||



