 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold02514-snap-gene-0.25-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 486aa MW: 53735.9 Da PI: 6.7868 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.5 | 2.1e-15 | 60 | 104 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rW++eE+++++da k++G W++I +++g ++ + q++s+ qk+
maker-scaffold02514-snap-gene-0.25-mRNA-1 60 RERWSEEEHKKFLDALKLYGRA-WRRIEEHVG-TKNAVQIRSHAQKF 104
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.92E-15 | 54 | 108 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.992 | 55 | 109 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 3.8E-15 | 58 | 107 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 7.3E-12 | 59 | 107 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-8 | 60 | 100 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.5E-12 | 60 | 103 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.10E-9 | 62 | 105 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 486 aa Download sequence Send to blast |
MAIQDQSVGS RSHSVVQASN GISLSSGLHS VTGVQLRDQF SCGNDYAPKV RKPYTITKQR 60 ERWSEEEHKK FLDALKLYGR AWRRIEEHVG TKNAVQIRSH AQKFFSKVAR DSNCSNTTST 120 EPIEIPPPRP KRKPMHPYPR KLVIPPNKEV SALEQPIRST SPNSSVWEQE DQSPTSVLSA 180 IGSDTVGFTD SDSPNGSLSP ASSTAGVQPE GLVLSEPNQS TEESASPSPA LKTVGSKFEL 240 FPRENNYVKE GASEETSNRS LKLFGRTVLV TDSHRPSSPT IVAGKSMPAD ILEEKVVQPL 300 PWNFVPLKST QGNTEYSWSP STQGGLYFMQ CQKENSYMAE ANSAAPIPWW TFYGGLRFPV 360 MPFHEVEQSK AYLGSDLEEI QGKEVQKEGS WTGSNNGSVD DGNNGDKCSD AETQSCPLFL 420 KEENQEAGLV FQLRPSENSA FSELGRSPGK CLKGFVPYKR CVAERHTQSS TIRGEEREEK 480 QIRLCL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
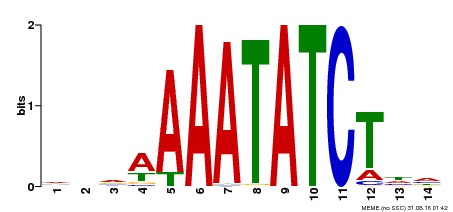
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023872232.1 | 0.0 | protein REVEILLE 1 | ||||
| TrEMBL | A0A2N9GSA3 | 0.0 | A0A2N9GSA3_FAGSY; Uncharacterized protein | ||||
| STRING | VIT_04s0079g00410.t01 | 1e-178 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2956 | 33 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 8e-67 | MYB_related family protein | ||||



