 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold02563-augustus-gene-0.16-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 431aa MW: 46416.3 Da PI: 5.8173 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 31.1 | 4.3e-10 | 273 | 321 | 3 | 55 |
HHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 3 rahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L+el+P + +k Ka +L + ++Y++sLq
maker-scaffold02563-augustus-gene-0.16-mRNA-1 273 NSHSLAERVRREKISERMRLLQELVPGC----NKITGKAVMLDEIINYVQSLQ 321
58**************************....999*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 6.8E-17 | 268 | 337 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.54E-11 | 268 | 325 | No hit | No description |
| PROSITE profile | PS50888 | 15.485 | 270 | 320 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-16 | 271 | 338 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.8E-7 | 273 | 321 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.6E-10 | 276 | 326 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009637 | Biological Process | response to blue light | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 431 aa Download sequence Send to blast |
MDGHENEDVG FQHGSEGILN CPSSRMSTNP LPEKVSAMTM SSVSMYKPSN GADPFFGSGW 60 DPLVSLSQSE NFGGSSMVSH GEYPPYPVVM ENQGISTTSH LVQYPSSSSF VELVPKLPCF 120 GSGSFSEMVN SFGLPDCSQI ANSGCPQNYA SNKEVATGRT STNGAQSKDD QQVSGEGAIG 180 SSPNAKRRKR VPDSNNSAFS PNQNGEGDVQ KDLSGDSSDG QKEPDEKKPK IEQNTGSNMR 240 GKQTSKQAKD TSSGDAPKDN YIHVRARRGQ ATNSHSLAER VRREKISERM RLLQELVPGC 300 NKITGKAVML DEIINYVQSL QQQVEFLSMK LATVNPELNI DIEQILSKDI IHSRGGTGAI 360 LGFGPGMSSS HPYPHGIYQG TLPSIPSTTP KYPSLSQNVL DNDFQSLFQM GFDSSSAIDN 420 LGPNGHLKPE N |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00135 | DAP | Transfer from AT1G10120 | Download |
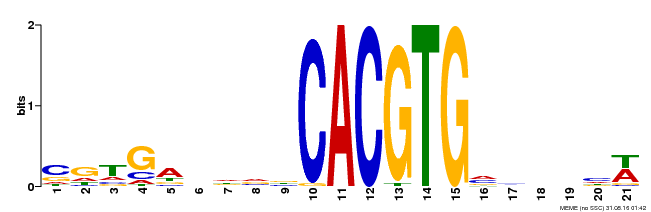
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023872654.1 | 0.0 | transcription factor bHLH74 | ||||
| TrEMBL | A0A2N9F2F5 | 0.0 | A0A2N9F2F5_FAGSY; Uncharacterized protein | ||||
| STRING | XP_008220406.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7585 | 33 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G10120.1 | 6e-84 | bHLH family protein | ||||



