 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold04781-augustus-gene-0.9-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 300aa MW: 34577.5 Da PI: 6.3476 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.5 | 2.7e-17 | 20 | 67 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd ll+++++ G g+W++ a++ g++Rt+k+c++rw++yl
maker-scaffold04781-augustus-gene-0.9-mRNA-1 20 RGPWTLEEDSLLIHYIAHNGEGRWNLLAKRSGLRRTGKSCRLRWLNYL 67
89********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48 | 2.9e-15 | 73 | 116 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ +++E++l+++++ ++G++ W++Ia++++ gRt++++k++w++
maker-scaffold04781-augustus-gene-0.9-mRNA-1 73 RGNLSPQEQLLILELHCKWGNR-WSKIAQHLP-GRTDNEIKNYWRT 116
7899******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 25.445 | 15 | 71 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.86E-29 | 18 | 114 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.2E-15 | 19 | 69 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.2E-16 | 20 | 67 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-21 | 21 | 74 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 9.69E-11 | 22 | 67 | No hit | No description |
| PROSITE profile | PS51294 | 19.245 | 72 | 122 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.5E-13 | 72 | 120 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-14 | 73 | 116 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-22 | 75 | 121 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 7.27E-10 | 77 | 116 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 300 aa Download sequence Send to blast |
MSTCSKSISS SSEDDSELRR GPWTLEEDSL LIHYIAHNGE GRWNLLAKRS GLRRTGKSCR 60 LRWLNYLKPD VKRGNLSPQE QLLILELHCK WGNRWSKIAQ HLPGRTDNEI KNYWRTRVQK 120 QARHLKIDTN SIAFQDIIRN YWMPRLIQKI EESSSSGVLF QNSAIPQPLD SAYQHSVTTA 180 PAQVVMQGPL CLSETIQSLD HHEQNPESES MNVLQIPQFS ECPTSQFHAK VNNDDGTFVK 240 DYYYADNNSY DVEAFNLESI SPLENFEDSS SKSHVVENNW ANNDFSGSMW NMDGLWQIRN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 4e-24 | 17 | 129 | 4 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00159 | DAP | Transfer from AT1G25340 | Download |
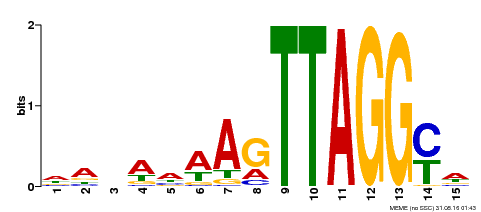
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023896380.1 | 0.0 | transcription factor MYB62-like | ||||
| TrEMBL | A0A2N9ES28 | 1e-161 | A0A2N9ES28_FAGSY; Uncharacterized protein | ||||
| STRING | XP_008240255.1 | 1e-129 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1639 | 34 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68320.1 | 1e-82 | myb domain protein 62 | ||||



