 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | maker-scaffold05269-snap-gene-0.14-mRNA-1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Fagaceae; Castanea
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 427aa MW: 46342.1 Da PI: 9.7578 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 43.8 | 5.7e-14 | 349 | 406 | 5 | 62 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkse 62
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L++ N++L+ + ee+ ++ ++ k e
maker-scaffold05269-snap-gene-0.14-mRNA-1 349 RRQRRMIKNRESAARSRARKQAYTLELEAEVAKLKEMNQELQRKQEEIMEMEKNQKLE 406
79***************************************99999999987766655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 7.7E-13 | 345 | 411 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.864 | 347 | 398 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 7.4E-13 | 349 | 398 | No hit | No description |
| CDD | cd14707 | 2.04E-26 | 349 | 403 | No hit | No description |
| Pfam | PF00170 | 5.0E-12 | 349 | 406 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 8.44E-10 | 349 | 398 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 352 | 367 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 427 aa Download sequence Send to blast |
MGSHINFKNF GDAPPGEGKP QGNYSLARQS SVYSLTFDEF QNTMGGFGKD FGSMNMDELL 60 KNIWTAEEIQ GMTCTVGGGE GSVHGGNLQR QGSLTLPRTI CQKTVDEVWK DLIKESSDGG 120 GGGAKVGNGN ANGVSNANVP QRQPTLGEVT LEEFLVRAGV VREDTQPTVR PNNSVFYGEL 180 PQPNNNTGLT FEFQQPNRSN GVLGNRIMEN NNSVPIQPPS LAMNVGGLRS SQQQIQQLQQ 240 QQSLFPKPTT VPFASPVHLV NNAQIANPGS RGSVVGVAEP SMNTALVQGG GIGRVGLAPG 300 VVTTIAGSPV SQISSEAKST IDTSSLSPVP YVFSRGRKCG GALEKVVERR QRRMIKNRES 360 AARSRARKQA YTLELEAEVA KLKEMNQELQ RKQEEIMEME KNQKLETMNR QWGGKRQCLR 420 RTLTGPW |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the ABA-responsive element (ABRE). Mediates stress-responsive ABA signaling. {ECO:0000269|PubMed:11884679, ECO:0000269|PubMed:15361142}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
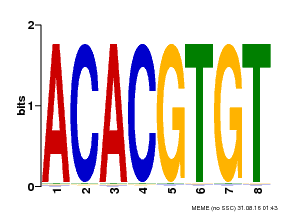
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA). {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:16284313}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023912796.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| Refseq | XP_023912797.1 | 0.0 | ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| Swissprot | Q9M7Q3 | 1e-114 | AI5L6_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ||||
| TrEMBL | A0A2I4EER7 | 0.0 | A0A2I4EER7_JUGRE; ABSCISIC ACID-INSENSITIVE 5-like protein 7 | ||||
| STRING | XP_008236589.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1962 | 34 | 81 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G19290.1 | 1e-98 | ABRE binding factor 4 | ||||



